
One of the sad, and unfortunate facts of life, is that many Anthropologists, Researchers and Academics, still refuse to acknowledge that Africans were the first Human colonizers of Europe, not to mention the rest of the world too. To hide this fact, they prefer to use the terms Aurignacian and Gravettian cultures to describe their habitations; which is fine, it still means Grimaldi. However, the current fad of attributing Grimaldi artifacts to the Humanoid hybrid Cro-Magnon is in all ways, mystifying. After all, thought different, Cro-Magnon like Grimaldi, was a Black African: albeit an admixture of Modern Humans and Neanderthal, so what's the point? Clearly obfuscation can be the only goal.
The Aurignacian culture is an archaeological culture of the Upper Palaeolithic, located in Europe and southwest Asia. It lasted broadly within the period from ca. 45,000 to 35,000 years ago The name originates from the type site of Aurignac in the Haute-Garonne area of France. The people of this culture also produced some of the earliest known cave art, such as the animal engravings at Aldène and the paintings at Chauvet cave in southern France. They also made pendants, bracelets and ivory beads, and three-dimensional figurines. Perforated rods, thought to be spear throwers or shaft wrenches, are also found at their sites.
The Gravettian toolmaking culture was a specific archaeological industry of the European Upper Palaeolithic era prevalent before the last glacial epoch. It is named after the type site of La Gravette in the Dordogne region of France where its characteristic tools were first found and studied. The earliest signs of the culture were found at Kozarnika, Bulgaria. One of the earliest artifacts is also found in eastern Crimea (Buran-Kaya) (see Crimean Mountains) dated 32,000 years ago. It lasted until 22,000 years ago. Where found, it succeeded the artifacts datable to the Aurignacian culture.
 |
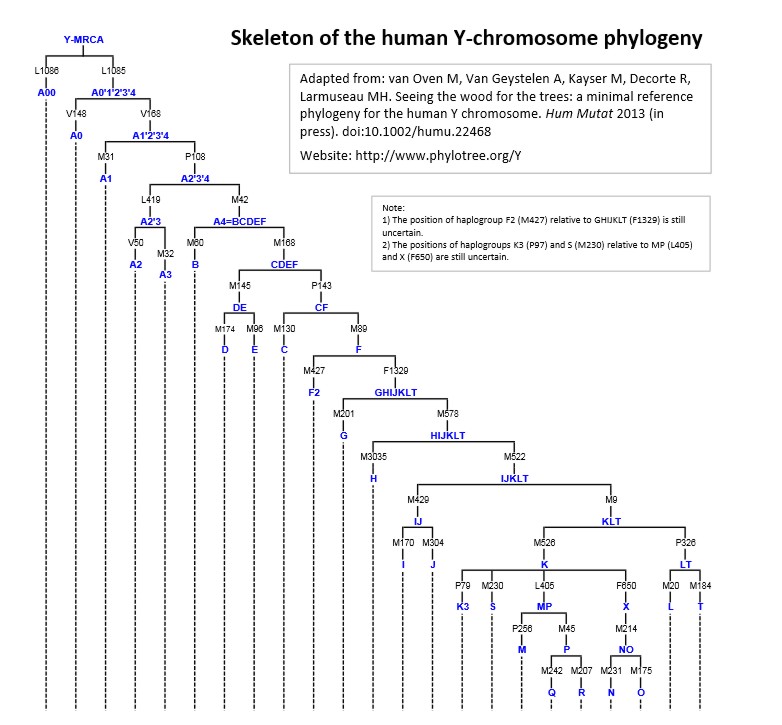
When investigations into human genetics first began, Albino scientists thought that it would prove their origins, their superiority, and clearly delineate between Whites and all other people. They even declared that Y-dna haplogroup "R" was unique to White/Albino people. Of course today, we know that Y-dna haplogroup "R" is actually an AFRICAN haplogroup and is found in many African locations. Later we found that it was not possible to determine "RACE" by DNA haplogroups. This is of course quite consistent with the reality that White people are merely the Albinos of Blacks, since Albinism does not effect the entire genetic makeup of a person, they retain their original (Black) genetic make-up.
Today, Albino scientists latest nonsense is the concept of Black Africans of the OOA migrations, upon once leaving Africa, experiencing genetic mutations causing a change in Haplogroup, and then RE-ENTERING Africa as the now "Mutated" human with the new haplogroup. That is how they try to explain haplogroups that are found widely in other parts of the world and Africa too.
 |
 |
In human mitochondrial genetics, Haplogroup M is a human mitochondrial DNA (mtDNA) haplogroup. An enormous haplogroup spanning all the continents, the macro-haplogroup M, like its sibling N, is a descendant of haplogroup L3.
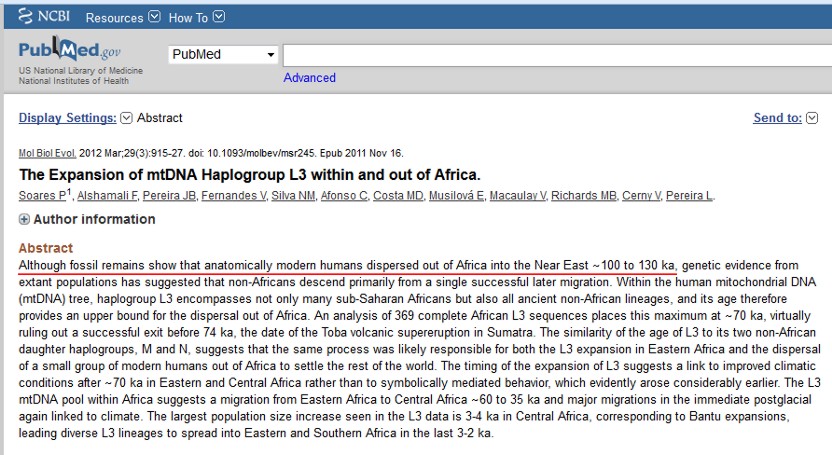
All mtDNA haplogroups considered native outside of Africa are descendants of either haplogroup M or its sibling haplogroup N.[9] The geographical distributions of M and N are associated with discussions concerning out of Africa migrations and the subsequent colonization of the rest of the world. In particular, it is often taken to indicate that it is very likely that there was one particularly major prehistoric migration of humans out of Africa, and that both M and N were part of this colonization process.
Haplogroup M1
Much of discussion concerning the origins of haplogroup M has been related to its subclade haplogroup M1, which is the only variant of macrohaplogroup M found in Africa. Two possibilities were being considered as potential explanations for the presence of M1 in Africa:
M was present in the ancient population which later gave rise to both M1 in Africa, and M more generally found in Eurasia.
The presence of M1 in Africa is the result of a back-migration from Asia which occurred sometime after the Out of Africa migration 40,000 years ago.
In human mitochondrial genetics, Haplogroup N is a human mitochondrial DNA haplogroup. An enormous haplogroup spanning many continents, the macro-haplogroup N, like its sibling M, is a descendant of haplogroup L3.
All mtDNA haplogroups found outside of Africa are descendants of either haplogroup N or its sibling haplogroup M. M and N are the signature haplogroups that define the out of Africa migration and the subsequent spread to rest of the world. The global distribution of haplogroups N and M, indicates that very likely, there was one particularly major prehistoric migration of humans out of Africa, and both N and M were part of the same colonization process.
 |
Haplogroup R is the main subclade of N, the one that was to generate the 6 most common European haplogroups (H, V, J, T, U, K). At the time of writing R subclades were numbered from R0 (a.k.a. pre-HV) to R31. Most of them are found in South Asia (R5, R6, R7, R8, R30, R31), Southeast Asia (R9, R21, R22, R24), East Asia (R9/F, R11/B), and even among Papuans (R14) and Australian aborigenes (R12). R0a peaks in the southern Arabian peninsula is common among Arabs and Middle-Easterners. R1a (not to be confused with the homonymous Y-chromosome haplogroup) is found among the Adygei people from the North Caucasus (related to the Maykop culture => see R1b section), Brahmins from northern India, northwestern Russians and Poles - basically all people closely related with the Indo-European expansion. R2 is found from northwest India and Pakistan to Iran, Georgia and Turkey. It could be connected to the Indo-Iranians.
Finno-Uralic mtDNA
Finno-Uralic people have an overall mtDNA admixture similar to other Europeans, with a higher percentage of W and U5b, and a small percentage of Siberian haplogroups such as N or A. The Sami are characterised by a high percentage of haplogroups U5b1 and V.
Berber mtDNA
The Berbers are the indigenous populationof north-west Africa. Although their Y-DNA is almost perfectly homogenous, belonging to haplogroup E-M81, Berber maternal lineages show a much greater diversity, as well as regional disparity. At least half (and up to 90% in some regions) of the Berbers belong to some Eurasian lineages, such as H, HV, R0, J, T, U, K, N1, N2, and X2, mostly of Middle or Near Eastern origin. 5 to 45% of the Berbers will have sub-Saharan mtDNA (L0, L1, L2, L3, L4, L5). There are only three native North African lineages, U6, X1 and M1, representing 0 to 35% of the people depending on the region.
Haplogroup U6 has been observed from the Iberia and the Canary Islands to Senegal in the West, and from Syria to Ethiopia and Kenya in the East. It is also found at low density in Europe, though mostly limited to Iberia. Approximately 10% of all North Africans belong to this lineage.
Gypsy mtDNA
The Gypsies (Romani people) originated in the Indian subcontinent and mixed with local population in the Middle East and Eastern Europe over the centuries. About half of the Gypsy population belong to haplogroup M, and more specifically M5 (reflected by Y-haplogroup H1a), which is otherwise exclusive to South Asia. The other mtDNA haplogroups found among the Gypsy community are mostly of Eastern European, Caucasian or Middle Eastern origin, such as H (H1, H2, H5, H9, H11, H20, among others), J (J1b, J1d, J2b), T, U3, U5b, I, W et X (X1b1, X2a1, X2f) (sources). The same diversity exist on the Y-DNA side (45% of H1a, followed by I1, I2a, J2a4b, E1b1b, R1b1b, R1a1a).
Saami and Berbers—An Unexpected Mitochondrial DNA Link
Abstract
The sequencing of entire human mitochondrial DNAs belonging to haplogroup U reveals that this clade arose shortly after the “out of Africa” exit and rapidly radiated into numerous regionally distinct subclades. Intriguingly, the Saami of Scandinavia and the Berbers of North Africa were found to share an extremely young branch, aged merely 9,000 years. This unexpected finding not only confirms that the Franco-Cantabrian refuge area of southwestern Europe was the source of late-glacial expansions of hunter-gatherers that repopulated northern Europe after the Last Glacial Maximum but also reveals a direct maternal link between those European hunter-gatherer populations and the Berbers.
Because of maternal transmission and lack of recombination, the sequence differentiation of human mtDNA has been generated by only the sequential accumulation of new mutations along radiating maternal lineages. Over the course of time, this process of molecular divergence has given rise to monophyletic units that are called “haplogroups.” Because this process of molecular differentiation occurred mainly during and after the process of human colonization of and diffusion into the different continents and regions, haplogroups and subhaplogroups tend to be restricted to specific geographic areas and population groups (Wallace 1995; Achilli et al. 2004).
Only the founders of the sister superhaplogroups M and N (which includes haplogroup R) (Quintana-Murci et al. 1999) participated in the “out of Africa” exit (Cann et al. 1987; Stringer and Andrews 1988; Cavalli-Sforza et al. 1994; Underhill et al. 2000; Forster et al. 2001) and were successful in colonizing the rest of the Old World. Superhaplogroup N is globally distributed outside Africa, encompassing virtually all of the western Eurasian mtDNA variation, and embraces haplogroup U, nested in haplogroup R. Haplogroup U has an extremely broad geographic distribution that ranges from Europe and North Africa to India and Central Asia and has a very high overall frequency (15%–30%) (Richards et al. 2000; Kivisild et al. 2003; Quintana-Murci et al. 2004)
To assess the nature and extent of haplogroup U variation, we initially sequenced 28 entire U mtDNAs from a wide range of populations. These were selected through a preliminary sequence analysis of the mtDNA control region to include the widest possible range of haplogroup U internal variation. A tree of the mtDNA sequences reveals that haplogroup U first splits into two major subsets, distinguished by the mutation at nt 1811, and that there is a very large number of independent basal branches. Among these, representatives of all known subhaplogroups (U1–U9) were included. However, subhaplogroup U5 provided a rather intriguing result. A Yakut from northeastern Siberia and a Fulbe from Senegal harbored mtDNAs that differed at only two coding-region nucleotide positions.
Haplogroup H is the most common and most diverse maternal lineage in Europe, in most of the Near East and in the Caucasus region. The Saami of Lapland are the only ethnic group in Europe who have low percentages of haplogroup H, varying from 0% to 7%. The frequency of haplogroup H in Europe usually ranges between 40% and 50%. The lowest frequencies are observed in Cyprus (31%), Finland (36%), Iceland (38%) as well as Belarus, Ukraine, Romania and Hungary (all 39%). The only region where H exceeds 50% of the population are Asturias (54%) and Galicia (58%) in northern Spain, and Wales (60%).
Haplogroup H possesses approximately 90 basal subclades identified to date, most of which further subdivided in other subclades. The most common subclades are H1, H2, H3, H4, H5, H6, H7, H10, H11, H13, H14 and H20. The Cambridge Reference Sequence (CRS), the human mitochondrial sequence to which all other sequences are compared, belongs to haplogroup H2a2a.
H1 & H3
Haplogroup H1 is by far the most common subclade in Europe, representing approximately than half of the H lineages in Western Europe. Roostalu et al. (2006) estimate that H1 arose around 22,500 years ago. H1 is divided in 65 basal subclades. The largest, H1c, has over 20 more basal subclades of its own, most with deeper ramifications. H1 is found throughout Europe, North Africa, the Levant, Anatolia, the Caucasus, and as far as Central Asia and Siberia. The highest frequencies of H1 are observed in the Iberian peninsula, south-west France and Sardinia. H3 has a very similar distribution to H1, but more confined to Europe and the Maghreb, and is generally two to three times less common than H1.
Haplogroup H5 is also found throughout western Eurasia and North Africa, but has a more Central European focus. Its highest frequencies are observed in Wales (8.5%), Slovenia (8%), Latvia (7%), Belgium (7%), Romania (6%), Bosnia-Herzegovina (5.5%), Russia (5%), Germany (5%), Slovakia (5%), Switzerland (4.5%) and Poland (4.5%).
Origins & History
The mutation defining haplogroup H took place at least 25,000 years ago, and perhaps closer to 30,000 years ago. Its place of origin is unknown, but it was probably somewhere around the northeastern Mediterraean (Balkans, Anatolia or Levant), possibly even in Italy.
Post-LGM recolonisation of Europe
During the Last Glacial Maximum (LGM, c. 26,500 to 19,000 ybp), human populations in Europe were mostly confined to refugia in southern France, Iberia, Italy, the Balkans and the Pontic Steppe. When the climate started warming up and the ice caps progressively receded, people recolonised Europe from these LGM refugia. Only a few partial ancient mtDNA sequences from this period have been tested. While those from Central and Eastern Europe consistently belonged to haplogroup U (U2, U5, U8), some samples from Spain, Portugal and Italy dating from 18,000 to 10,500 years ago could have belonged to haplogroup H, and possibly even H1 or H3 (although the partial results cannot confirm this with 100% certainty). The modern distributions of H1 and H3 support a Late Glacial recolonisation of Western and Central Europe from the Franco-Cantabrian refugium.
Some people have argued that H1 and H3 only came with Neolithic farmers from the Near East. The oldest incontrovertible evidence of the presence of H1 and H3 in Europe comes from the 5,000-year-old site of Treilles in Languedoc, France. 17 mtDNA sequences were retrieved from this Cardium Pottery culture site, including three that belonged to haplogroup H1 and three to H3. The men belonged to the Near Eastern Y-DNA haplogroup G2a and the Mesolithic European haplogroup I2a. It is noteworthy though that the I2a men belonged exclusively to H1 and H3, a potent, albeit not conclusive evidence that H1 and H3 were lineages that belonged to Mesolithic hunter-gatherers from south-west Europe.
Other H subclades were also probably found among Mesolithic or Late Upper Paleolithic Europeans based on their exclusive presence in Europe today. This could be the case of haplogroups H10, H11, H17, H45, as well as many minor subclades for which too little data is available at the moment, but that seem to be exclusively European. H10 and H11 have a stronger presence in Eastern and Central Europe and would have re-expanded from the northern Black Sea region rather than from Southwest Europe after the LGM.
Neolithic diffusion of agriculture
The development of the Neolithic lifestyle in the Fertile Crescent was about to bring a wave of Near Eastern immigrants that would change dramatically the genetic landscape of Europe. Goats and sheep were the first animals domesticated, in the Taurus and Zagros mountains some 11,000 years ago. The domestication of wild cereals could have taken place as early as 23,000 years before present in the southern Levant, although the actual cultivation of wheat and barley didn't start until 11,500 ybp. The first pottery produced in the Fertile Crescent appeared circa 6500 BCE. It is from this period that early agriculturalists began expanding towards western Anatolia and Greece. These Neolithic farmers potentially belonged to haplogroups H2, H5, H7, H13 and H20 (as well as J2b1, K1a, N1, T2 and X2), all of which have been found in ancient Neolithic samples from Europe and are also found throughout the Middle East today.
One group migrated by sea to Italy and to the western Mediterranean, creating the Cardium Pottery culture. The H subclades most closely associated with this culture is H13, which would have been a lineage of goat herders living in mountainous environment. Nowadays H13 is one of the most common H subclades in the Caucasus (13% in Georgia, 15% in Daghestan) and in Sardinia (9%), two regions with high levels of Y-haplogroup G2a, J1 and J2a. H13 is also relatively common (2 to 8%) all the way from the Levant to Iberia, but rare in northern Europe.
Another Neolithic expansion started off from the Balkans, commencing in Thessaly circa 6500 BCE and spreading at first (6300-5800 BCE) to modern Albania, Macedonia, Bulgaria and Serbia. It is likely that this Thessalian Neolithic was conducted not only by Near Eastern immigrants (Y-haplogroups G2a, E-M123, J1 and T), but also by assimilated local populations (Y-haplogroups E-V13, J2b and maybe I2a1 too). From 5500 to 4500 BCE, the Linear Pottery culture (LBK) spread along the Danube from northern Serbia to Germany and all the way to the Netherlands and Poland. These farmers might also have taken brides among the neighouring tribes of hunter-gatherers, as suggested by the small amount of Mesolithic European admixture in a 7,500-year old individual from the LBK culture. Over 100 mtDNA samples from this period have been tested and included haplogroups H2, H5, H7 and H20, among the likely newcomers from the Middle East, but also H1, H3, H10, H11a, H16 and H89, among the probable lineages coming from assimilated Mesolithic European populations.
H7 was probably part of the Neolithic migration from the Carpathians to Ukraine that gave rise to the Dnieper-Donets culture, along perhaps with mt-haplogroups K1c, K2b, T1a1a, T2a1b1 and T2b, and with Y-haplogroups G2a3b1 and J2b2. All these lineages would subsequently be absorbed by the Proto-Indo-European speakers (Y-haplogroups R1a and R1b) of the Yamna culture during Bronze Age.
Neolithic & Chalcolithic Europe
H1 and H3 lineages would have been some of the most prevalent mt-haplogroups among the Megalithic cultures of Western Europe, which spanned the whole Neolithic and Chalcolithic periods, from the 5th millennium BCE until the arrival of the Proto-Celts (Y-DNA R1b) from 2200 BCE to 1800 BCE (or up to 1200 BCE in parts of Iberia). Megalithic people would have belonged essentially to Y-haplogroups I2, G2a and E1b1b, with the possible addition of J2 lineages during the Chalcolithic. From the Bronze Age, R1b male lineages replaced a large percentage of Megalithic Y-haplogroups, but female Megalithic lineages survived almost unchanged in frequency. Celtic culture was born from the fusion of Indo-European paternal lineages (R1b) with native Central and Western European maternal lineages (including H1, H3, H10, J1c, K1a, T2, U5 and X2).
The presence of H1 was confirmed in remains from the Late Neolithic Funnel Beaker culture in Scandinavia, which can also be classified as a Megalithic culture.
Indo-European invasions during the Bronze Age
The Bronze Age brought on a new wave of immigrants from the East that would have a tremendous impact on the gene pool and cultural identity of Europe: the Proto-Indo-European (PIE) people. Originating in the Pontic-Caspian Steppe, to the north of the Black Sea and the Caucasus, PIE speakers were the first people to use bronze technologies for military purposes, starting from c. 3700 BCE. They were also the first to domesticate horses, which they supposedly rode to manage their cattle in the extensive steppes. Already living a (semi-)nomadic existence as cattle herders since the Neolithic (=> see R1b history), horse riding and bronze working provided PIE steppe people with the means to invade and conquer practically any part of the world they wished. They started with Europe, invading the Carpathians and the Balkans repeatedly between 4200 and 2800 BCE, before eventually leaving the steppes almost completely and moving further west across Europe, until the Atlantic coast. The northern branch of PIE speakers, associated with the development of Balto-Slavic and Indo-Iranian languages (=> see R1a), invaded Poland, Germany and Scandinavia to the west from 2900 to 2400 BCE, and Siberia and Central Asia to the east, from 2500 to 1800 BCE.
PIE speakers from the Pontic Steppe are known to have possessed quite different maternal lineages from that of Mesolithic and Neolithic Europeans. A few H subclades linked to the R1a populations of Northeast Europe would obviously have been found among Mesolithic Eastern Europeans, like H1b, H1c and H11. Others may have come from the North Caucasus, like H2a1, or from the advance of Neolithic farmers from the Balkans to Northeast Europe, like H7. Others yet would have come from Anatolia when R1b cattle herders crossed the Caucasus, attracted by the vast pastures of the Pontic Steppe. These would have included H5a, H8 and H15.
H4 and H6, both absent from Europe before the Bronze Age, have such a wide distribution across the continent nowadays that they would likely have been spread both by R1a and R1b branches of the Indo-Europeans.
The Corded Ware culture, which is associated with the expansion of Y-haplogroup R1a from the steppes to Central Europe and Scandinavia, yielded samples belonging to H1ca1, H2a1, H4a1, H6a1a and H10e. Ancient DNA from the Catacomb culture, strongly associated with Y-haplogroup R1a, yielded samples belonging to H1, H2a1 and H6. The Unetice culture, which is thought to mark the arrival of R1b in Central Europe (but overlapping with the previous R1a expansion), had individuals belonging to H2a1a3, H3, H4a1a1a2, H7h, H11a, H82a.
Remains from the Andronovo culture in Central Asia, which is associated with R1a people, only yielded a single H sample (H6).
Middle Eastern subclades
Some H subclades are rare in Europe and geographically confined mostly to the Middle East. This includes H14 and H18.
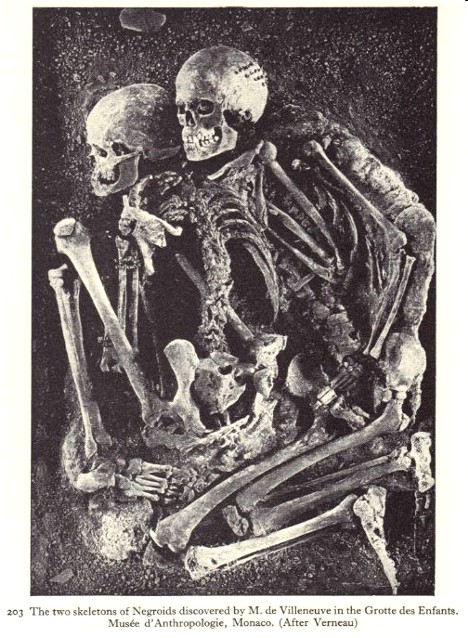 |
One of the first evidences of this historic journey of Grimaldi, was found at the caverns of Grimaldi (Baousse-Rousse), between Mentone and Ventimiglia and on the Italian side of the international boundary, these caverns form one of the most compact groups of paleolithic caverns in all Europe. Counting two small rock-shelters, the group includes nine stations, the most important being the Grotte des Enfants, La Barma Grande, Grotte du Cavillon and the Grotte du Prince. General attention was first called to this region many years ago by Dr. Paul Riviere's discovery of a human skeleton in the Grotte du Cavillon, the so-called homme de Menton, now in the Natural History Museum, Paris. Here Rivière demonstrated the presence of deliberate burials and ornamented clothing in 1872.
Later five skeletons in all were found at La Barma Grande, and two of children, in the Grotte des Enfants, whence its name. Interest in archeology and ownership of one of the caverns (Grotte du Prince), led the Prince of Monaco, Prince Albert I Grimaldi (1848-1922), to provide for a systematic exploration of the caverns: thus the fossils became known as "Grimaldi" in his honor. Fossil remains of these ancient Africans have also been found in France, Switzerland, Central Europe, Bulgaria, Russia, and as Far East as Siberia. The African Khoisan nature of the Fossils was first documented by Boule, Marcellin & Vallois, in their book "Fossil Men" The Dryden Press (1957).
 |
 |
 |
Just as exciting as the fossil finds, were the artifact finds at the Balzi Rossi, or Red Rocks (also known as the Grimaldi Caves), were Louis Alexandre Jullien, carried out the excavations that led, at the end of the last century, to the discovery of fifteen figurines, called the Balzi Rossi Venuses. Many other sites across Europe and Asia have also produced Grimaldi Venuses: many of them Steatopygia Females, and many of normal proportion females. Notable of these are the Venus of Willendorf: found near Krems, Austria. The Venus of Brassempouy, found in France in 1892. It should be noted; that though all Grimaldi figures are not of Steatopygia Females, that style of figure, is the signature of Grimaldi.
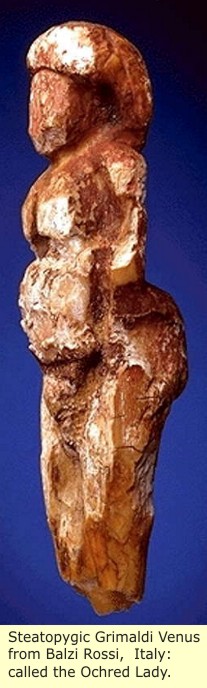 |
 |
 |
 |
 |
 |
One of the most exciting Grimaldi finds of the 19th century, was made by a Spanish nobleman: Sr. Don Marcelino de Sautoula, on his estate on the northern coast of Spain. An amateur prehistorian, he partially excavated a cave on a hill called Altamira. He found stag, horse, bison bones, oyster shells, numerous implements and bone tools characteristic of the Magdalenian culture. Exploring deeper into the cave, he encountered cave bear bones and extensive ashes. On one of his excavation trips, he took his 12-year-old daughter, Maria, with him into the cave. While he sat digging in damp darkness under the light of a lantern, the girl suddenly cried out, "Papa, papa. Mira. Toros pintados." (Father, father, Look Painted bulls.) Maria showed her astonished father that the roof of the cavern was covered with a superb fresco of red and black bison. Lantern light gave them three-dimensional form since the artist had incorporated the natural projections of the ceiling with great skill into his art. Sautuola was deeply moved as he and his daughter explored the cavern roof and saw marvelous polychrome animals, executed in three-dimensional skill, all painted by an artist who had known the animals before they became extinct from Spain in the distant past.
The first Homo sapiens (Grimaldi man) arrived by small groups in northern Spain around 45,000 - 35,000 BP. They cohabited for a time with the last of the Neanderthals, and then developed a significant culture known as paleolithic cave art which developed across Europe, from the Urals to the Iberian Peninusula, from 35,000 to 11,000 BC. Because of their deep galleries, isolated from external climatic influences, these caves are particularly well preserved. The caves are inscribed as masterpieces of creative genius and as the humanity’s earliest accomplished art.
The last Ice Age then began to make its influence felt, ending in around 18,000 BP. During this period cave art developed in the eastern part of Cantabria, producing an individual style (Altamira, La Peña del Candamo, El Castillo, Las Pasiega, El Pendo, La Garma, Chufin and El Pendo).
The artistic apogee, known as Magdalenian, corresponds to the end of the Ice Age, from 17,000 to 13,000 BP. This was the period of the major works in the decorated caves, with a great variety of motifs and techniques of representation. This was one of the key moments of the history of art, as seen for example in the polychrome figures of Altamira and El Castillo, the combination of engraving and painting, the use of the rock forms themselves, and realistic detail in the animal figures in most of the nominated caves.
From 13,000 to 10,000 BP, the climate became warmer (Holocene), causing a profound transformation in human lifestyles, together with a decline in cave art. Las Monedas is an example of late cave art, and there is no evidence of cave art later than 11,000 BP.
The Chauvet-Pont-d'Arc Cave in the Ardèche department of southern France is a cave that contains some of the earliest known cave paintings, as well as other evidence of Upper Paleolithic life. It is located near the commune of Vallon-Pont-d'Arc on a limestone cliff above the former bed of the Ardèche River, in the Gorges de l'Ardèche. Discovered on December 18, 1994, it is considered one of the most significant prehistoric art sites. The dates have been a matter of dispute but a study published in 2012 supports placing the art in the Aurignacian period, approximately 30,000–32,000 BP. Based on radiocarbon dating, the cave appears to have been used by humans during two distinct periods: the Aurignacian and the Gravettian. Most of the artwork dates to the earlier, Aurignacian, era (30,000 to 32,000 years ago). The later Gravettian occupation, which occurred 25,000 to 27,000 years ago, left little but a child's footprints and other trivial things.
 |
 |
 |
Altamira is a cave in Spain famous for its Upper Paleolithic cave paintings featuring drawings and polychrome rock paintings of wild mammals and human hands. Its special relevance comes from the fact that it was the first cave in which prehistoric cave paintings had been discovered. When the discovery was first made public in 1880, it led to a bitter public controversy between experts which continued into the early 20th century, as many of them did not believe prehistoric man had the intellectual capacity to produce any kind of artistic expression. The acknowledgement of the authenticity of the paintings, which finally came in 1902, changed forever the perception of prehistoric human beings. It is located near the town of Santillana del Mar in Cantabria, Spain, 30 km west of the city of Santander.
Archaeological excavations in the cave floor found rich deposits of artifacts from the Upper Solutrean (c. 18,500 years ago) and Lower Magdalenean (between c. 16,500 and 14,000 years ago). Both periods belong to the Paleolithic or Old Stone Age. In the millennia between these two occupations, the cave was evidently inhabited only by wild animals.
 |
 |
 |
Lascaux is a complex of caves in southwestern France famous for its Paleolithic cave paintings. The original caves are located near the village of Montignac, in the department of Dordogne. They contain some of the best-known Upper Paleolithic art. These paintings are estimated to be 17,300 years old. They primarily consist of images of large animals, most of which are known from fossil evidence to have lived in the area at the time.
 |
 |
 |
The Cosquer cave is located in the Calanque de Morgiou in Marseille, France, not very far from Cap Morgiou. The entrance to the cave is located 37 m (121 ft) underwater, due to the rise of the Mediterranean in Paleolithic times. It was discovered by diver Henri Cosquer in 1985, but its contents were not made public until 1991, when three divers became lost in the cave and died. Today, the cave can be accessed by divers through a 175 m (574 ft) long tunnel, the entrance of which is located 37 m (121 ft) below the surface of the sea, because of changes in sea level since the time the cave was inhabited. The shore of the Mediterranean sea at the time the cave was occupied was then several km away and many metres below the cave mouth. Sea level was lower because at that time there was an ice age and large volumes of water were retained in enormous icecaps on land, making the level of the sea 110 to 120 m (360 to 390 ft) lower than today.
Four-fifths of the cave, including any art on its walls, was submerged and obliterated by the rising sea. 150 instances of cave art remain including several dozen painting and carvings dating back to the Upper Paleolithic, corresponding to two different phases of occupation of the cave: Older drawings consisting of 65 hand stencils and other related motifs, dating to 27,000 years BP (the Gravettian Era).
Newer drawings of signs and 177 animal drawings dating to 19,000 years BP (the Solutrean Era), representing both "classical" animals such as bison, ibex, and horses, but also marine animals such as seals and what appear to be auks and jellyfish.
 |
 |
 |
 |
 |
 |
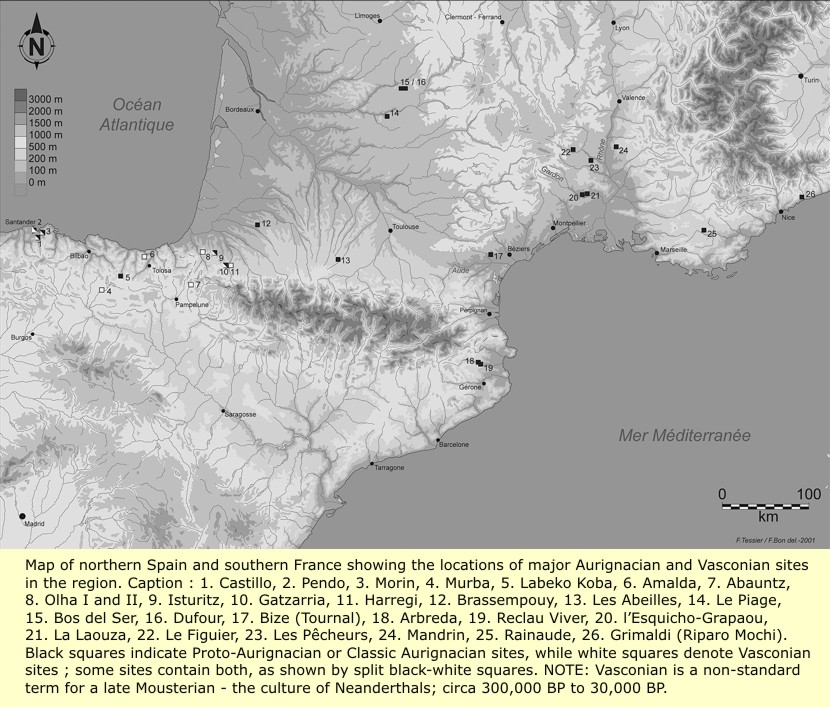 |
Total listing of the main Aurignacian sites in Northern Spain and Southern France, (Some of which are mixed sites). 1) Hornos dela Pena. 2) Castillo. 3) Pendo. 4) Morín. 5) Salitre. 6) Otero. 7) Polvorin. 8) Santimamine. 9) Ekain. 10) Labeko Koba. 11) Lezetxiki. 12) Aitzbitarte. 13) Chabiague. 14) Le Basté. 15) Isturitz. 16) Gatzarria. 17) Tercis, Moulin deBénesse. 18) Brassempouy. 19) Garet. 20) Cazaubon (Drouilhet). 21) Gargas. 22) Les Abeilles. 23) Aurignac. 24) Tarté. 25) Mas d’Azil. 26) Tuto deCamalhot. 27) Canecaude I. 28) Les Cauneilles-Basses. 29) Bize (Tournal). 30) Régismont-le--Haut. 31) La Crouzade. 32) Romani. 33) Cal Coix. 34) Can Crispins. 35) Bruguera. 36) Arbreda. 37) Mollet I. 38) Reclau Viver. 39) Rothschild. 40) L’Esquicho--Grapaou. 41) La Laouza. 42) La Balauzière. 43) La Salpêtrière. 44) Le Figuier, Les Pêcheurs. 47) Ségalar. 48) La Moulinière. 49) Beauville (Huiet Toulousète). 50) Las Pélénos. 51) Les Ardailloux. 52) Laburlade. 53) Abri Peyrony. 54) Le Piage. 55) Roc de Combe. 56) Les Fieux. 57) Laussel. 58) Le Flageolet, grotte XVI. 59) Caminade. 60) La Ferrassie. 61) La Faurélie. 62) Lartet et Poisson. 63) Pataud. 64) Cro-Magnon. 65) La Rochette. 66) Cellier. 67) Le Facteur. 68) Vallon deCastelmerle (abrisCastanet, Blanchard, La Souquette). 69) Labattut. 70) Belcayre. 71) La Bombetterie. 72) Bos del Ser. 73) Dufour. 74) Font-Yves. 75) Bassaler-Nord. 76) Chanlat. 77) Comba delBouïtou. 78) Barbas. 79) Champ-Parel. 80) Pair-non-Pair. 81) Roc deMarcamps. 82) Rochecourbon. 83) Gros Roc. 84) Grotte à Melon. 85) Combe deRolland. 86) Les Rois. 87) Les Vachons. 88) La Quina. 89) La Chaise.
The archaeological site known as Tianyuan Cave (Tianyuandong or Tianyuan 1 Cave) is located at the Tianyuan Tree Farm in Huangshandian Village, Fangshan County, China. Tianyuan Cave's opening is at 175 meters (575 feet) above current sea levels, higher than other sites at Zhoukoudian. The cave includes a total of four geological layers, only one of which--Layer III--contained human remains, the partial skeleton of an archaic human being. Numerous fragmentary evidence of animal bone have also been recovered, most notably in the first and third layers.
Although the context of the human bone was somewhat disturbed by the workmen who discovered the site, scientific excavations uncovered additional human bone in situ. The human bone has been interpreted to most likely represent Early Modern Human. The bones were radiocarbon-dated to between 42,000 and 39,000 calibrated years before the present. With that, the Tianyuan Cave individual is one of the oldest Early Modern Human skeletons recovered in eastern Eurasia, and in fact, is one of the earliest outside of Africa.
 |
Thirty-four human bones were removed from the cave, probably from a single individual of about 40-50 years of age, including a jaw bone, fingers and toes, both leg bones (femur and tibia), both scapulae, and both arm bones (both humeri, one ulna). The gender of the skeleton is indeterminate, since there was no pelvis recovered and long bone length and gracility measures are ambiguous. No skull was recovered; and neither were any cultural artifacts, such as stone tools or evidence of butchering on the animal bone. The age of the individual was estimated based on tooth wear and evidence for moderately advanced osteoarthritis in the hands.
The nuclear DNA sequences determined from this early modern human (Mtdna haplogroup "B") revealed that the Tianyuan individual derived from a population that was ancestral to many present-day Asians and Native Americans but postdated the divergence of Asians from Europeans. They also show that this individual carried proportions of DNA variants derived from archaic humans similar to present-day people in mainland Asia.
Originally published in Science Express on October 8 2015
www2.zoo.cam.ac.uk/manica/ms/2015_Gallego_Llorente_et_al_Science.pdf
Abstract
Characterizing genetic diversity in Africa is a crucial step for most analyses reconstructing the evolutionary history of anatomically modern humans. However, historic migrations from Eurasia into Africa have affected many contemporary populations, confounding inferences. Here, we present a 12.5x coverage ancient genome of an Ethiopian male (‘Mota’) who lived approximately 4,500 years ago. We use this genome to demonstrate that the Eurasian backflow into Africa came from a population closely related to Early Neolithic farmers, who had colonized Europe 4,000 years earlier. The extent of this backflow was much greater than previously reported, reaching all the way to Central, West and Southern Africa, affecting even populations such as Yoruba and Mbuti, previously thought to be relatively unadmixed, who harbor 6-7% Eurasian ancestry.
In 2011, archaeologists working with Gamo tribesman in the highlands of southwest Ethiopia discovered Mota Cave, 14 metres wide and 9 metres high, overlooking a nearby river. A year later, they excavated a burial of an adult male, his body extended and hands folded below his chin. Radiocarbon dating suggested that the man died around 4,500 years ago — before the proposed time of the Eurasian migrations and the advent of agriculture in eastern Africa.
Advances in ancient DNA technology allow researchers to reap DNA from ever older bones, and the cool, constant temperatures of caves are kind to the molecule. So a team co-led by Ron Pinhasi, an archaeologist at University College Dublin, tested the Mota man's bones for intact DNA and found enough to sequence his genome 12 times over.
The first-ever DNA sequencing of the skull of the ancient African – the 4,500-year old ‘Mota man’ has revealed that a huge migration from Western Eurasia into the Horn of Africa 3,000 years ago, was twice as significant in terms of numbers and genetic influence as had been thought. Indeed, it was so large that it could have increased the population of the Horn of Africa by close to a third – which in turn led to a bigger genetic impact than expected, the report found.
The man’s DNA suggests that Middle Eastern farmers migrated into Africa several thousand years ago, leaving traces of their Eurasian ancestry in the genomes of many modern-day Africans.
The man's genome is, unsurprisingly, more closely related to present-day Ethiopian highlanders known as the Ari than to any other population the team examined, suggesting a clear line of descent for the Ari from ancient human populations living in the area. But further genetic studies show that the Ari also descend from people that lived outside Africa, which chimes with a previous study that discovered a ‘backflow’ of humans into Africa from Eurasia around 3,000 years ago. (Humans first migrated FROM Africa some 60,000 to 100,000 years ago.)
Using genetic evidence from Eurasian ancient genomes and present-day populations, the researchers determined that the migrant ancestors of the Ari were closely related to early farmers who moved into Europe from the Near East around 9,000 years ago. Co-author Marcos Gallego Llorente, an evolutionary geneticist at the University of Cambridge, UK, suggests that Middle Eastern farmers later moved south to Africa, bringing new crops to the continent such as wheat, barley and lentils. The team also found vestiges of these migrants’ DNA in people all across sub-Saharan Africa — probably carried by later migrations, such as the expansion of Bantu-speaking groups from West Africa to other parts of the continent around 1,000 years ago.
Mota Man was assigned to Mtdna haplogroup L3x2a, and Y-haplogroup E1b1.
Supplementary Text has dna haplotypes
http://www.sciencemag.org/content/early/2015/10/07/science.aad2879/suppl/DC1
Comment: Basically what these researchers are saying is that Mota man shows no genetic commonality with Eurasians, but many MODERN Africans do. Therefore they are taking Mota man as being TYPICAL of Ancient Africans. And they are saying that the part of the modern Ari genome that is different from Mota man is due to Eurasian admixture.
There are a few problems with the assumptions and logic's used.
1) Africa is a HUGE place with well over a BILLION people, many in isolated communities - Researchers know little about African genetics. Yet these idiots on the basis of ONE sample, are presuming to know the genetics of ALL ancient Africans. This in the face of knowing that Africans have the worlds most varied genetics.
2) Then there is the problem of the Eurasian base samples, what skeletal remains were they taken from?
BECAUSE - NONE OF THE PEOPLE IN THE MIDDLE EAST ARE THE ORIGINAL POPULATIONS!
THEY ARE ALL TURKS AND TURK MULATTOES FROM THE ARAB AND TURKISH CONQUEST - FROM 640 A.D. ONWARD: (The Muslim conquest).
In the Europe and the Mediterranean, the modern people are the descendants of invading Central Asian Albinos: so the SOURCE genetic material becomes extremely important.
To be clear, European and Middle-Eastern Blacks, including Egyptians, did return to lower Africa. One needs only to look at the Hebrew Lemba in Zimbabwe and the Hebrews in Ethiopia to know that. It is the concept that this was a huge, Africa changing backflow, and is based on such flimsy evidence, that is absurd.
 |
 |
But playing Devils advocate and going along with this nonsense: These are some possible historical events that might have spurred Black Europeans and Middle-Easterners back into lower Africa 3,000 years ago.
1) The Egyptian invasion of the "Sea People": who were Black Europeans and Mediterranean's fleeing the FIRST invasion of the Central Asian Albinos - (these Albinos were finally incorporated into the Black Greek and Roman populations). Herodotus explains it thusly:
Herodotus Book 1 - CLIO
[1.58] The Hellenic race has never, since its first origin, changed its speech. This at least seems evident to me. It was a branch of the Pelasgic, which separated from the main body, and at first was scanty in numbers and of little power; but it gradually spread and increased to a multitude of nations, chiefly by the voluntary entrance into its ranks of numerous tribes of barbarians. The Pelasgi, on the other hand, were, as I think, a barbarian race which never greatly multiplied.
The texts and relief's that deal with the Sea Peoples date to year eight of Ramesess III’s reign, approximately 1190 B.C.
1a) The second wave of Central Asian Albinos started to arrive just before the modern era; these included the various Germanic tribes (Angles, Saxons, Vandals, Ostrogoths, Visigoths, Alans, Burgundians, Lombards, etc. And myriad Slav tribes, too numerous to count. These are the progenitors of modern Europeans.
2) The Sea People finally settled in Anatolia (modern Turkey), bringing an end to one of the Hattie nations (falsely called the Hittites). These supplanted Anatolian's may have returned to Africa.
3) By the mid-9th century B.C, two new groups of people appear in Elam (modern Iran), these are the Medes (Mada) and the Persians (Parsua). The two formed the Persian nation and gradually supplanted the Elamites. This might have caused supplanted Elamites to return to Africa.
For those familiar with the workings of the Albino mind: none can't help but wonder:
What could the Albinos possibly be up to with this silly study? Imagine, a study suggesting that Africa's Billion plus, could have been materially altered by a few Africans RETURNING to Africa. And then it struck me!
Remember when the Albinos were trying to say that EAST AFRICANS WEREN'T REALLY BLACKS! But really BLACK CAUCASIANS - BLACK WHITES! Well, I think that with this study, they are going to say that this BACKFLOW from EURASIA is what gave East Africans their Caucasian features!
Ha,ha,ha,ha: Obviously they don't know, or don't know that we know, EXACTLY what ancient Eurasians looked like! Please see the Canaan (Modern Palestine-Israel-Lebanon) section: The Elam (Modern Iran) section: The Sumer (modern Iraq) section: Along with the Minoan (Greece and the Mediterranean Islands) section: The Etruria (Rome and Italy) sections. They will learn that you cannot fool an educated people.
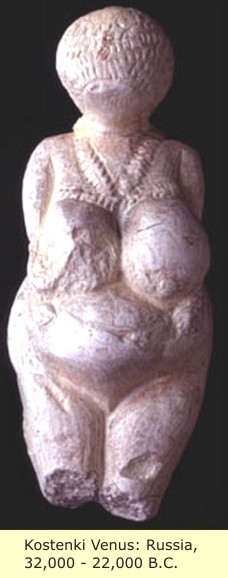
The Kostenki - Borshevo sites (34,000 B.C.) are a group of more than twenty settlements from the same culture, on the right bank of the Don River, south of Voronezh. The basic excavations were conducted in the 1920s - 1930s by P. Yefimenko, and in the 1940s - 1960s by A. N. Rogachev.
 |
The villages of Kostenki and Borshevo contained five cultural layers. In the upper layer were preserved the remains of dwellings with hearths located along the central longitudinal axis of the dwellings, together with storage pits. Flint tools and hoes made from mammoth tusks, bone digging implements, a baton from deer horn, and about forty female statuettes made from both ivory and marl/limestone, figurines of a bear, cavelion and anthropomorphous marl heads. Triangular flint tools are found in the lowermost layer with a concave base, retouched with a pressure process. At Kostenki II (Zamyatnina site) were found the remains of a round dwelling made of mammoth bones, seven or eight metres across, with the fireplace in the center. Kostenki man was found to be Male Y-dna = C-V199, Mtdna = U2.
The site of Sungir (26,000 B.C.), discovered during clay extraction operations in 1956, was excavated by Otto Bader from 1956 to 1977. Excavations were re-opened by Bader's assistant Ludmilla Mikhailova and Bader's son Nicolai in 1986, and continue today. Sungir is an enormous early Upper Paleolithic living site located on the outskirts of the city of Vladimir, 192 km from Moscow in the Russian Republic.
While inhabiting Sungir, at least five of the site's occupants perished. According to Russian physical anthropologists, these consisted of a 60 year-old man, a 7 to 9 year-old girl, a 13 year-old boy, an unsexed headless adult and an adult female skull. The two adolescents and the adult male were buried in two shallow graves three metres apart, dug into the permafrost beneath the living surface of the site. All three of the corpses were laid on their backs with their hands folded across their pelvises. The fourth individual was represented by an isolated and poorly preserved female skull placed beside a stone slab in an area stained with red ochre, and was found overlying the man's burial (a person sacrificed to serve as a protector in the afterlife?). The fifth skeleton, that of a headless adult, was so poorly preserved as to be practically unrecoverable. It was found immediately on top of the two adolescents, (a person sacrificed to serve as a protector in the afterlife?), who were buried together in a head-to-head fashion in the middle of an apparently abandoned circular dwelling structure.
 |
 |
 |
Each of the three intact individuals were lavishly decorated with thousands of painstakingly prepared ivory beads arranged in dozens of strands, perhaps basted to their clothing. Although it is almost certain that the three individuals buried intact at Sungir were members of the same social group, there are remarkable differences among them in details of body decoration and grave offerings. The man was adorned with 2,936 beads and fragments arranged in strands found on all parts of his body including his head, which was apparently covered with a beaded cap that also bore several fox teeth. His forearms and biceps were each decorated with a series of polished mammoth-ivory bracelets (25 in all), some showing traces of black paint. They were thin, flat strips of mammoth-ivory, cut longitudinally along the tusk. They were pierced at each end, some with one hole, others with two, apparently to keep the ivory bent into a circle. What appear to be brush strokes from the application of pigment are visible on at least one specimen. Around the man's neck, he wore a small, flat schist pendant, painted red, but with a small black dot on one side. (DNA testing on the Sungir remains were done, but the Albino scientists have so far refused to divulge the results. No doubt because it is another major blow against the false histories of the Albino people). New - Sungir Mtdna = H2a2a, but termed unreliable.
Paglicci Cave is an archaeological site situated in Paglicci, near Rignano Garganico, Apulia, southern Italy. The cave, discovered in the 1950s. In the cave, situated near Rignano Garganico, there are more than 45,000 individual finds, including Paleolithic tools, human and animal bones. They are currently housed in Rignano Garganico's Museum.
The cave contains also some Paleolithic mural paintings, depicting horses and handprints. Images of goats, cows, a serpent, a nest with eggs, and a hunting scene have also been found engraved on bone. Two human skeletons have been found as well, belonging to a boy and a young woman, both wearing deer bone or teeth ornaments. Paglicci cave contains the earliest Aurignacian and Gravettian remains of Italy, dated to c. 34,000 and 28,000 BP. In 2008, a scientific team led by David Caramelli tested human remains from Paglicci cave (Paglicci 23) dated 28,000 BP and found that the individual had the human mitochondrial haplogroup "H".
 |
 |
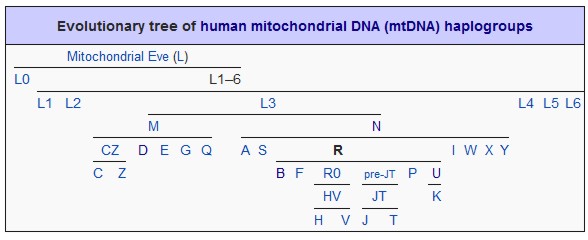
The base mtDNA test is the HVR-1 test. The HVR-1 region contains an abundance of SNP markers and is useful for maternal lineage searches. The HVR-2 region contains additonal markers which will increase the resolution of the search. The Coding region also contains SNP markers which will proivide even further enhancement of resolution once a match is found.
Testing the SNP markers in all 3 regions is required to confirm an individual's mtdna Haplogroup and Subclade. Since the HVR1 and HVR2 regions are relatively short (approximately 500 base pairs in length), sequencing technology can be used to read all of the markers in the entire HVR1 and HVR2 regions. However, the Coding Region of the mtDNA is extremely long, containing over 15,000 base pairs, thus making sequencing very difficult as many sequencing reactions are required to read all of the markers in the entire coding region.
Once your mtDNA HVR1 sequence is known, it is compared to a reference sequence known as the “Cambridge Reference Sequence” (CRS). Any region of your sequence that differs from the CRS is considered a SNP mutation.
The Cambridge Reference Sequence (CRS) for human mitochondrial DNA was first published in 1981 leading to the initiation of the human genome project. A group under Dr. Fred Sanger at Cambridge University sequenced the mitochondrial genome of one individual of European descent during the 1970s, determining it to have a length of 16,569 base pairs (0.0006% of the total human genome) containing some 37 genes. The reference sequence belongs to European haplogroup H2a2a. The revised CRS is designated as rCRS.
 |
 |
Paglicci site Southern Italy
(Paglicci 23), dated 28,000 years BP Mtdna haplogroup H.
(Paglicci-12), dated 24,000 years BP Mtdna haplogroup N*
(Paglicci 25), dated 23,000 years BP Mtdna haplogroup HV.
Kostenki Russia, dated 34,000 BP Mtdna haplogroup U2.
Grimaldi at Mal’ta Siberia, dated 24,000 BP Mtdna haplogroup U, Y-dna R.
Ötzi the Iceman, northern Italy 5,300 years BP. Mtdna haplogroup K, Y-dna G2a2b.
King Tutankhamun of Egypt, Y-dna haplogroup R1b1a2. His Mtdna haplogroup is still secret.
King Ramesses III of Egypt, 3,200 BP Y-dna haplogroup E1b1a, of East Africa Origin, a YDNA haplogroup that predominates in most Sub-Saharan Africans. His Mtdna haplogroup is still secret.
Etruscan Mtdna
Etruscan DNA from tombs of the 7th. century to the 3rd. century B.C. in Etruria. Around 13 are H, others possibly J/T. In the new Etruscan studies, they refuse to divulge the haplogroups, that is a good indicator that the results prove African ancestry.
A European population in Minoan Bronze Age Crete (Racist)
Jeffery R. Hughey, Peristera Paschou, Petros Drineas, et al, 14 May 2013.
Using the DNAs of 37 Minoans whose remains were well preserved in a cave ossuary located in the Lassithi plateau of east-central Crete. The majority of Minoans were classified in haplogroups H (43.2%), T (18.9%), K (16.2%) and I (8.1%). Haplogroups U5A, W, J2, U, X and J were each identified in a single individual.
Quote: Our calculations of genetic distances, haplotype sharing and principal component analysis (PCA) exclude a North African origin of the Minoans. Instead, we find that the highest genetic affinity of the Minoans is with Neolithic and modern European populations. We conclude that the most likely origin of the Minoans is the Neolithic population that migrated to Europe about 9,000 YBP.
Quote from analysis: A clear issue is that the current inhabitants of the plateau have a distinctive genetic signature in their Y-DNA, quite different from that of other Cretans, with much higher frequencies of R1b and R1a and much much lower frequencies of the most common Cretan lineage: J2a1. However they also almost lack the main mainland Greek haplogroup E1b, what suggests that the recolonization from Peloponnese story is not correct either.
Interestingly Cretan R1b, so important in Lasithi Plateau (almost 50%), is also largely derived from Western Europe (although the other half could be Balcanic), maybe via Italy, and cannot be ancestral to it (almost all the Western variant belongs to a derived subclade common in Italy, Central Europe and France: U152).
http://www.nature.com/ncomms/journal/v4/n5/full/ncomms2871.html
Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau (Racist)
The most outstanding differences were observed in haplogroups J2 and R1, with the predominance of haplogroup R lineages in the Lasithi Plateau and of haplogroup J lineages in the more accessible regions of the island. Y-STR-based analyses demonstrated the close affinity that R1a1 chromosomes from the Lasithi Plateau shared with those from the Balkans, but not with those from lowland eastern Crete. In contrast, Cretan R1b microsatellite-defined haplotypes displayed more resemblance to those from Northeast Italy than to those from Turkey and the Balkans.
http://www.nature.com/ejhg/journal/v15/n4/full/5201769a.html
Byzantine Mtdna
Mitochondrial analysis of a Byzantine population By analysing the mtDNA variation in 85 skeletons from Sagalassos dated to the 11th–13th century A.D. Claudio Ottoni, François-X Ricaut, Ronny Decorte, et al, May 2011.
All Sagalassos sequences fall into a set of haplogroups within macrohaplogroups N (X, W and N1b) and R (R0a, H, V, HV, U, K, J and T) that are characteristic of West Eurasians.
Mitochondrial DNA in Ancient Human Populations of Europe - Doctoral thesis by Clio Der Sarkissian July 2011.
In combination with coalescent age calculations, the founder
analysis identified sub-clades likely to have reached Europe during the initial
colonisation of Europe during the Upper Palaeolithic (haplogroup U in the Early
Upper Palaeolithic: ~45,000 – 55,000 yBP, haplogroups HV, I and U4 in the Middle
Upper Palaeolithic: ~17,000 - 38,000 yBP, haplogroups H, K and T2 in the Late Upper
Palaeolithic:~8,000 -17,000 yBP), and those likely to derive from the genetic input of
Neolithic migrants from the Near East (haplogroups J1a and T, ~6,000 – 13,000 yBP;
Richards et al., 2000). Based on these estimates, Richards et al., 2000 concluded that
~80% of the present-day European mtDNA had arisen from Upper-Palaeolithic
hunter-gatherers versus ~20% from migrating Near East farmers of the Neolithic
(Richards et al., 2000).
In order to reconstruct human population dynamics in north east Europe,
we examined mitochondrial DNA (mtDNA) from four diachronically sampled
archaeological sites: two Mesolithic sites at Uznyi Oleni Ostrov and Popovo (n = 9
and n = 2; 7,500 years before present, yBP), a Bronze Age site at Bolshoy Oleni
Ostrov (n = 23; 3,550 yBP) and a historical Saami graveyard Chalmny-Varre (n = 42,
200 – 300 yBP). Important temporal information emerges from the new genetic data
obtained from 76 human remains and suggests a complex genetic history in north east
Europe. Prehistoric foragers - found at Uznyi Oleni Ostrov and Popovo - showed high
frequencies and diversity in haplogroups U4 and U5a, a pattern previously observed in
populations of ancient European hunter-gatherers. The Bronze Age population of
Bolshoy Oleni Ostrov was clearly differentiated from Mesolithic hunter-gatherers due
to the presence of mtDNA haplogroups C, D and Z of eastern Siberian origin.
Historical Chalmny-Varre was found to be closely linked to modern-day Saami due to
the presence of haplogroup V and the ‘Saami motif’ U5b1b1.
The Mitochondrial Gene Pool of Scythians of the Rostov Area, Russia:
Mitochondrial haplogroup structure of the Scythians
and comparison with
modern-day populations of Eurasia
Mitochondrial haplogroups detected in Scythians were distributed into
‘western’ and ‘eastern’ haplogroups as follows: 62.4% ‘western’ (12.4% H, 12.4% I,
18.9% T, 6.3% U2, 12.4% U5), 31.3% ‘eastern’ (6.3% A, 6.3% C, 12.4% D, 6.3% F)
and 6.3% ‘other’ (U7). Distributions of ‘western’ and ‘eastern’ lineages similar to the
Scythian mtDNA makeup were found in modern populations of Bukharan Arabs and
Khoremians of Uzbekistan, both located in Central Asia, as well as in Udmurts of east
Europe (Figure 1C; Table 3). When analysed with PCA, the mtDNA haplogroup
frequencies observed in Scythians (SCY) fell close to modern-day Europeans on the
PCA biplots (Figure 2). Scythians were found at the periphery of the homogeneous
cluster of western/central Europeans, close to populations of eastern Europeans such
as Tatars (TA2), Pomors (pom), Udmurts (UD) of western Russia, as well as
Bulgarians (BGR) and Albanians (ALB) of the Balkans. The position of the Scythians
on the biplot was also close to present-day Central Asian Shugnans of Tajikistan (shu),
which are characterised by a high frequency of ‘western Eurasian’ haplogroups, in
particular H (29.4%).
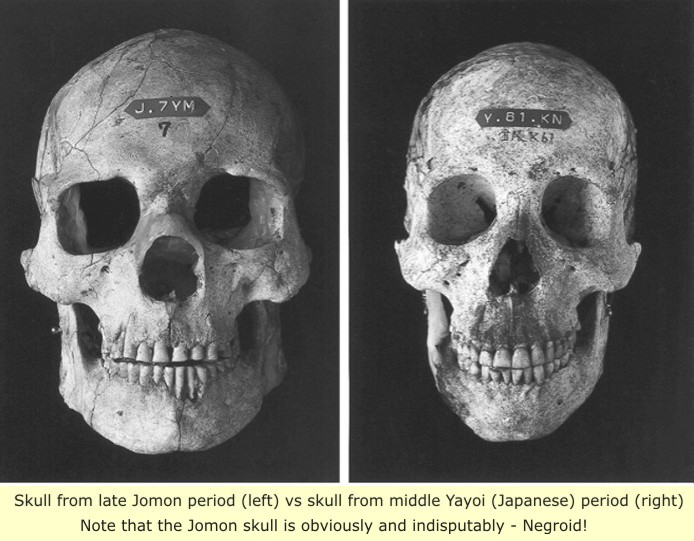
We identified haplogroups from all four samples that were analyzed; haplogroup frequencies were 50% (n = 2) for N9b and 50% (n = 2) for M7a2. Haplogroup N9b has been previously observed in high frequencies in the other Tohoku Jomon, Hokkaido Jomon, Okhotsk, and Ainu peoples, whereas its frequency was reported to be low in the Kanto Jomon and the modern mainland Japanese. Sub-haplogroup M7a2 has previously been reported in the Hokkaido Jomon, Okhotsk, and modern Udegey (southern Siberia) peoples, but not in the Kanto Jomon, Ainu, or Ryukyuan peoples. Principal component analysis and phylogenetic network analysis revealed that, based on haplogroup frequencies, the Tohoku Jomon was genetically closer to the Hokkaido Jomon and Udegey people, than to the Kanto Jomon or mainland modern Japanese.
At present, 70 Jomon-period skeletons have been tested in two separate studies (although both in Hokkaido only). The haplogroups identified were D1, D4h2, G1b, M7a, N9b. The dominant lineage was by far N9b. Haplogroup N9b is unique to Japan and is the maternal equivalent of C1 and D2 on the paternal side. N9a is found in Southeast Asia, parts of China, and throughout Japan, but is absent from Korea or Eastern China, and is consequently also surely of Jomon origin. M7, although present in most of East Asia, is almost always represented by the M7a subclade in Japan, and has been associated with the Jomon and Ainu people.
Y-DNA haplogroup C3 and N permeated the Jomon stock. Y-haplogroup C, which has been associated with the first migration of modern humans out of Africa towards Asia, is relatively frequent in Kerala (southern tip of India) and Borneo. These early Austronesians are thought to have been the ancestors of the Ice Age settlers of Japan.
Study - Global distribution of Y-chromosome haplogroup C reveals the prehistoric migration routes of African exodus and early settlement in East Asia.
Zhong, Shi, et al. May 2010.
 |
Friedlander et. al. 2005, 2007
Melanesian populations Y-dna (haplogroups F, G,H, K and C). These Melanesian haplogroups, found in an increasing cline from ISEA to New Guinea and Near Oceania, still bear the Y-SNP signature of the first Paleolithic settlers who initially crossed ISEA, coming directly from Africa and following the southern coastal route.
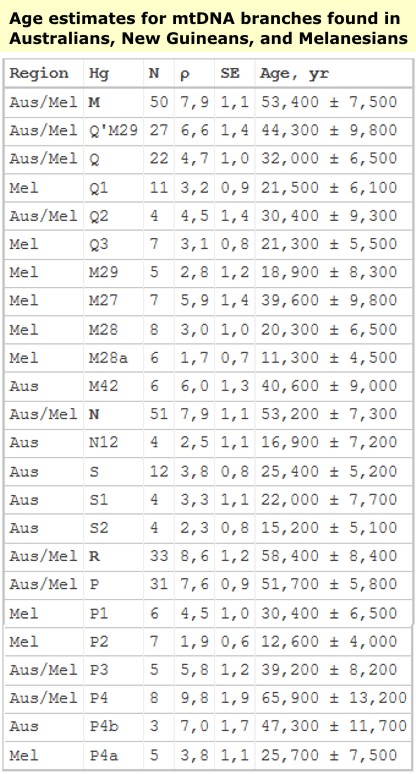
MtDNA - All extant Asian or Melanesian individual mtDNA types are descendent of founding macro haplogroupeither type M or type N. These two mtDNA haplogroupsshare a common ancestry with African super haplogroupL3 which was carried by the only small group of people who successfully passed through the horn of Africa~80 000 YBP and later migrated out of Africa ~60,000 YBP as a group bearing only haplogroups M and N (the two daughters of haplogroup L3). In less than 5,000 years (a time that was too short to allow for the appearance or the fixation of new mutations of mtDNA macro haplogroups N or M), these peoples established settlements in India, Sundaland (MSEA), Papua New Guinea and Australia. Much later, in the last 15,000 years when circumstances, dictated by fluctuations in sea levels and climatic conditions, were more favourable they settled in America. Interestingly, European ancestors from west Eurasia (a small group of people all belonging to mtDNA haplogroup N) moved to Europe much later than the eastern wave (~35 000 YBP) and intrinsically the genetic diversity observed in Europeans is less than the genetic diversity seen in Asians which itself is less than the diversity of Africans.
Solomon Islands
All the retrieved ancient Solomon Islands sequences belong to East Asian, southeast Asian (B4/B4a, B4a1a1) and ancient Near Oceanic (Q1, M27) mtDNA haplogroups, common amongst populations from Island Melanesia Friedlaender et al. 2007 Despite the small size of our 19th-century Western Solomon Islands sample, the mtDNA genepool composition is relatively similar to that of modern Solomon Islanders.
Y-dna Haplogroup C & D
Haplogroup C seems to have come into existence shortly after M168 was introduced, probably at least 60,000 years before present. Although Haplogroup C attains its highest frequencies among the indigenous populations of Mongolia, the Russian Far East, Polynesia, Australia, and at moderate frequency in the Korean Peninsula and Manchuria, it displays its highest diversity among modern populations of India, and therefore it is hypothesized that Haplogroup C either originated or underwent its longest period of evolution and diversification within India or the greater South Asian coastal region.
It represents a great coastal migration along Southern Asia, into Southeast Asia and Australia, and up the Asian coast. It is believed to have migrated to the Americas some 6,000-8,000 years before present, and was carried by Na-Dené speaking peoples into the northwest Pacific coast of America ( Dene or Dine (the Athabaskan languages) is a widely distributed group of Native languages spoken by associated peoples in Alberta, British Columbia, Manitoba, Northwest Territories, Nunavut, Saskatchewan, Yukon, Alaska, parts of Oregon, northern California, and the American Southwest as far as northern Mexico). Some have hypothesized that Haplogroups C and D were brought together to East Asia by a single population that became the first successful modern human colonizers of that region, but at present the distributions of Haplogroups C and D are different, with various subtypes of Haplogroup C being found at high frequency among the Australian aborigines, Polynesians, Vietnamese, Kazakhs, Mongolians, Manchurians, Koreans, and indigenous inhabitants of the Russian Far East and at moderate frequencies elsewhere throughout Asia and Oceania, including India and Southeast Asia, whereas Haplogroup D is found at high frequencies only among the Tibetans, Japanese peoples, and Andaman Islanders, and has been found neither in India nor among the aboriginal inhabitants of the Americas or Oceania.
| Note: Polynesian is a term that the Albino people have applied to Pacificans/Austronesians who have significant "White Mongol/European" admixture. They reserve the term Melanesian for the original "Pure Black" Pacificans/Austronesians who have resisted admixture. |
Philippines
Aeta people: Y-dna haplogroups C, K, NO, O.
Mitochondrial DNA Haplogroups from Four Negrito Populations two Aeta, two Agta.
B4a1a, D4, M7b3, M7c3c, Y2, B4b1, B5b, F1a3a, F1a4a, E1b1, R9*/F*, P9, P10.
Andaman Islands
Y-dna: Male Onges and Jarawas almost exclusively belong to Haplogroup D-M174.
Y-dna: Male Great Andamanese have an assortment of other paternal haplogroups besides D-M174, these clades include O, L, K and P-M45.
Mtdna: All Andamanese belong to the subgroup M (M2 and M4).
 |
 |
 |
The vast territory of North and Central Asia represents a poorly understood region in the prehistoric era, despite intensive excavations that have been conducted during the past century, the earliest human occupation in this region probably began sometime around 40,000 years ago. Small groups of big-game hunters likely migrated into this region from lands to the south and southwest, confronting a harsh climate and long, dry winters, by about 22,000 BP, two principal cultural traditions had developed in Siberia and northeastern Asia: the Mal'ta and the Afontova Gora-Oshurkovo - which is mostly about tools.
Modern human (i.e., Homo sapiens sapiens) remains from the sites of Afontova Gora II and Mal'ta contain the first evidence of Mongoloid features in Siberia, circa 21,000 years BP.
The Mal'ta tradition is known from a vast area spanning west of Lake Baikal and the Yenisey River. The site of Mal'ta is composed of a series of semi-subterranean houses made of large animal bones and huge numbers of reindeer antlers, (no doubt scavenged for hut construction after being shed by the reindeer). The huts had likely been covered with animal skins and sod to protect the inhabitants from the severe, prevailing northerly winds bearing loess dust from the edges of the glaciated regions. Among the artistic accomplishments evident at Mal'ta are remains of expertly carved bone, ivory, and antler objects. Figurines of birds and human females are the most commonly found items.
Evidence seems to indicate that Mal'ta is the most ancient site in eastern Siberia, however relative dating illustrates some irregularities. The use of flint flaking and the absence of pressure flaking used in the manufacture of tools, as well as the continued use of earlier forms of tools seem to confirm the fact that the site belongs to the early Upper Paleolithic. Yet, it lacks typical skreblos (large side scrapers,) that are common in other Siberian Paleolithic sites) Additionally, other common characteristics such as pebble cores, wedge-shaped cores, burins, and composite tools have never been found) The lack of these features, combined with an art style found in only one other nearby site, make Mal'ta culture unique in Siberia.
 |
Twenty nine ivory female figures were found on the open site of Mal'ta. They differ from contemporary representations in Russia, Central and Western Europe in that they are shown clothed rather than nude, most have faces and the body shapes are straight or tapering below large, round heads. Some are also perforated to be worn as pendants.
 |
 |
Close-up |
 |
 |
 |
In late 2009, researchers sampled at the Hermitage Museum, St. Petersburg Russia: the remains of a boy from the Upper Palaeolithic site of Mal’ta in Siberia. The juvenile dated to approximately 24,000 years ago. Mal’ta, located along the Belaya River near Lake Baikal, was excavated between 1928 and 1958 and yielded a plethora of archaeological finds including 30 anthropomorphic Venus figurines, which are rare for Siberia but found at a number of Upper Palaeolithic sites across western Eurasia.
A double burial of two children was identified at Mal'ta, covered by a stone slab. The oldest individual was a single Homo sapiens boy of about 3-4 years old (MA-1), wearing a necklace of beads, several pendants and an ivory diadem. Partial remains include parts of the cranium, mandible and maxilla, and several post-cranial bones. The second child is only represented by teeth. Both have slightly shovel-shaped incisors, a characteristic of North American and modern Siberian people, but according to the Nature paper (Raghavan et al. 2013), bioarchaeologist Christy Turner investigated and concluded that the bones are morphologically most closely related to Upper Paleolithic Europeans. Other grave goods interred with the children include a decorated plaque, a bird-shaped pendant, an ivory bracelet, stone tools and an ivory baton.
“Representing the oldest anatomically modern human genome reported thus far, the MA-1 individual has provided us with a unique window into the genetic landscape of Siberia some 24,000 years ago“, says Dr. Maanasa Raghavan from the Centre for GeoGenetics and one of the lead authors of the study. “Interestingly, the MA-1 individual shows little to no genetic affinity to modern populations from the region from where he originated – south Siberia.” Instead, both the mitochondrial and nuclear genomes of the boy indicate that he was related to modern-day western Eurasians. This result paints a picture of Eurasia 24,000 years ago which is quite different from the present-day context. The genome of the boy indicates that prehistoric populations related to modern western Eurasians occupied a wider geographical range into northeast Eurasia than they do today.
One of the researchers, Dr. Willerslev, an expert in analyzing ancient DNA, was seeking to understand the peopling of the Americas by searching for possible source populations in Siberia. He extracted DNA from bone taken from the child’s upper arm, hoping to find ancestry in the East Asian peoples from whom Native Americans are known to be descended. But the first results were disappointing.
Pending further testing, Willerslev assigned the Mal'ta sample to hg U (mtDNA) and hg R (Y-DNA). This is the earliest attestation of these haplogroups in Siberia suggesting that these West Eurasian lineages were much more widely distributed in Eurasia in pre-LGM times and common in Eastern Eurasia, too. The results are consistent with the finding of Mtdna haplogroup "U2" in 34,000-year-old Kostenki remains in Central Russia, and support the pattern whereby the earliest ancient DNA samples (Tianyuan Cave in China with hg "B", Kostenki with hg "U2" and now Mal'ta with hg "U" have so far turned up members of mtDNA macro-haplogroup R. While the dates of all these samples are consistent with 50,000 YBP chronological frame proposed by geneticists for the divergence of mhg R, it's still noteworthy that not only is the most downstream mtDNA macrohaplogroup also the most widely spread among human populations, but also that it seems to be more wide-spread in the past than now.
The first research on living Native American tribes showed they were composed of four distinct mtDNA haplogroups called A, B, C, and D which means that the Native Americans are derived from four different lineages. These haplogroups were also found in native populations in Central and South America. Other mtDNA research utilizing ancient remains recovered in the Americas validated these four haplogroups. Three of these haplogroups, A, C, and D are found primarily in Siberian Asia. The B haplogroup, however, is found only in aboriginal groups in Southeast Asia, China, Japan, Melanesia, and Polynesia.
Based on the mutations found in the mtDNA, most researchers think that groups A, C and D, entered America from Siberia across Beringia some time around 35.000 B.C. Group B, they assert, probably came to America from the South Pacific or Japan via boats. It is believed the B groups began this migration not long after the A, C, and D groups arrived. However, the majority of the B group arrived about 11,000 B.C. This leaves open the possibility of several migrations by the B group from different locations. It should be noted that a few geneticists have proposed that each of these haplogroups came in four separate migrations while many Clovis supporters argue that all the groups migrated together.
NativeAmericanCouncilIn 1997, a fifth mtDNA haplogroup was identified in Native Americans. This group, called X is present in three percent of living Native Americans. Haplogroup X was not found in Asia, but was found only in Europe and the Middle East where two to four percent of the population carry it. In those areas, the X haplogroup has primarily been found in parts of Spain, Bulgaria, Finland, Italy, and Israel. A few people with the X type have been identified in the Altasians tribe located in extreme southern Siberia in the Gobi Desert area. In addition, the 'X’ type has now been found in the ancient remains of the Basque people.
Archaeologists and geneticists are certain that the presence of X in America is of ancient origin and not the result of historic intermarriages. Among Native American tribes, the X haplogroup has been found in small numbers in the Yakima, Sioux, and Navaho tribes. It has been found to a larger degree in the Ojibway, Oneonta, and Nuu-Chah-Nulth tribes. The X haplogroup has also been discovered in ancient remains in Illinois near Ohio and a few areas near the Great Lakes. It has not yet been found in South or Central American tribes including the Maya. The X haplogroup appears to have entered America in limited numbers perhaps as long ago as 34,000 B.C. appearing in much greater numbers from 12,000 B.C. to 10,000 B.C.
Three Amerindian populations, two from Rondônia, Brazil (Karitiana and Rondônia Suruí), and one from Campeche, Mexico (Mayan) were DNA typed, the results are not available.
Polynesian culture refers to the indigenous peoples' culture of Polynesia who share common traits in language, customs and society. Recent maternal mitochondrial DNA analysis suggests that Polynesians, including Samoans, Tongans, Niueans, Cook Islanders, Tahitians, Hawaiians, Marquesans and Māori, are genetically linked to indigenous peoples of parts of Southeast Asia including those of Taiwanese aborigines. This DNA evidence is supported by linguistic and archaeological evidence. Recent studies into paternal Y chromosome analysis shows that Polynesians are also genetically linked to peoples of Melanesia. However the "out of Taiwan model" has been recently challenged by a study from Leeds University and published in Molecular Biology and Evolution. Examination of mitochondrial DNA lineages shows that they have been evolving within Island Southeast Asia (ISEA) for a longer period than previously believed. Polynesians arrived in the Bismarck Archipelago of Papua New Guinea at between 3500 and 3000 years ago and modern Polynesians are the result of a few Austronesian seafarers mixing with Melanesians. The population migrations were most likely to have been driven by climate change — the effects of the drowning of a huge ancient peninsula called ‘Sundaland’ (that extended the Asian landmass as far as Borneo and Java). This happened during the period 15,000 to 7,000 years ago following the last Ice Age. Oppenheimer outlines how rising sea levels in three massive pulses caused flooding and the submergence of the Sunda Peninsula, creating the Java and South China Seas and the thousands of islands that make up Indonesia and the Philippines today.
In 2007, the skeleton of a teenage girl was found in an underwater cave in Mexico. The new discovery comes from a spectacular underwater cave on Mexico’s Yucatán Peninsula. Divers stumbled across a trove of prehistoric animal bones and one of the oldest, most complete human skeletons in the Western Hemisphere. The human remains are those of a teenage girl, who apparently took a fatal tumble into the limestone cavern between 12,000 and 13,000 years ago, before slowly rising seas engulfed the formation. The divers who found the skeleton in 2007 named her “Naia,” Greek for water nymph.
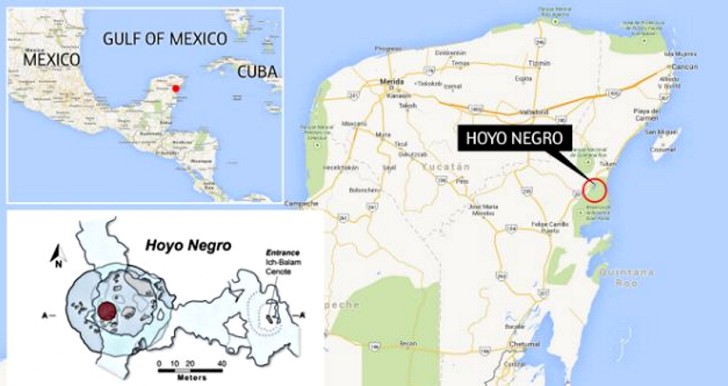 |
Alberto Nava, one of the divers who discovered the skeleton in 2007, said he and his colleagues swam through a mile-long tunnel before reaching a chamber the size of a basketball arena. They named it Hoyo Negro, or Black Hole, because it was so vast their underwater lights barely penetrated the darkness. Heaps of bones on the chamber floor, nearly 150 feet below sea level, later were identified as those of sabertooth cats, elephant-like creatures called gomphotheres, and giant sloths. Naia’s bones were lying on a small ledge. The girl probably fell through a sinkhole into the chamber, which at that time was above sea level, Chatters said. Her pelvis was broken. “I think she died almost instantly, if not instantly,” he said.
 |
 |
 |
Jim Chatters, as the first scientist to study the skeleton of Kennewick Man, unearthed in Eastern Washington almost two decades ago. Chatters was embroiled in a controversy over race and cultural identity stirred up by the 9,500-year-old bones. His assertion that the mystery man didn’t look anything like modern Native Americans infuriated Northwest Tribes, who consider the remains those of an ancestor and sued for the right to rebury what they call the Ancient One. Now the Bothell archaeologist is back in the spotlight with another set of prehistoric bones, along with DNA evidence that helps resolve a long-standing puzzle about the first Americans.
 |
The young woman’s skeleton shares many of the physical traits that led Chatters to question Kennewick Man’s relationship to modern Native Americans. Quote: “Even though she is extremely feminine looking, and he is very masculine looking, they look a lot alike,” he said. Both skeletons have narrow brain cases, short faces and prominent foreheads typical of people from the Pacific Rim, Australia and Africa.
 |
Native Americans more closely resemble (MODERN) people from northeast Asia. That jibes with genetic studies documenting their descent from Siberians believed to have migrated east into the land mass that once linked Asia and Alaska, and thence into the Americas beginning about 17,000 years ago. To explain why the bones of the Western Hemisphere’s oldest inhabitants — called paleoamericans — have such an unexpected appearance, Chatters and other scientists hypothesized that the Americas were colonized twice in prehistoric times: first by people from Southeast Asia or even Europe, then by migrants from Siberia.
From DNA extracted from Naia’s teeth (mtdna D1), Genetic analyses conducted at Washington State University and other labs show a clear link between the girl in the cave and modern Native Americans. The study is the first to show that despite having unusual features, at least one paleoamerican — Naia — is descended from the same ancestors as modern Native Americans, said WSU anthropologist Brian Kemp.
After a lengthy custody battle with Northwest tribes and federal agencies, a group of scientists led by Smithsonian anthropologist Doug Owsley was allowed to study the Kennewick Man bones in 2005. Owsley, who declined to comment on the new report, said his team’s book-length manuscript will be published this fall. Owsley is steadfast in his belief, based on physical features, that the ancient man is not genetically linked to the tribes who still hope to lay the bones to rest someday.
Scientists would love to extract DNA from Kennewick Man, but the agencies who control access to the skeleton won’t allow it. Without those results, it’s impossible to know if Kennewick Man would share Naia’s genetic affinity with Native Americans. But the results from Mexico add ammunition to the tribes’ argument that just because his head is shaped differently from theirs, doesn’t mean they aren’t related, Kemp said. For nearly 20 years, since Kennewick Man turned up, I’ve been wondering why these early people looked so different from Native Americans,” Chatters said, this is one step toward resolving that issue.” New - Kennewick = Y-dna = Q-M199, Mtdna = X2a
Naia’s skull shape does not look like those of Native Americans, but the Beringian-derived mitochondrial DNA D1 haplogroup directly links her to the modern Native peoples of the Americas. It is found in Northeast Asia (including Siberia). Its subclade D1 (along with D2, D3, and D4h3a) is one of five haplogroups found in the indigenous peoples of the Americas, the others being A, B, C, and X. Haplogroup D is also found quite frequently in Central Asia, where it makes up the second most common mtDNA clade (after H). Haplogroup D also appears at a low frequency in eastern Europe and southwestern Asia. There are two principal branches, D4 and D5'6. D1 is a basal branch of D4 that is widespread and diverse in the Americas. D2, which occurs with high frequency in some arctic and subarctic populations (especially Aleuts), is a subclade of D4e1 parallel to D4e1a and D4e1c, so it properly should be termed D4e1b. D3, which has been found mainly in some Siberian populations and in Inuit of Canada and Greenland, is a branch of D4b1c.
D4 (3010, 8414, 14668): The subclade D4 is the most frequently occurring mtDNA haplogroup among modern populations of northern East Asia, such as Japanese, Okinawans, Koreans, and Mongolic or Tungusic-speaking populations of northern China. D4 is also the most common haplogroup among the Buryats and Khamnigans of the Buryat Republic, the Kalmyks of the Kalmyk Republic, and the Telenghits and Kazakhs of the Altai Republic. It is spread also all over China, Southeast Asia, Siberia, Central Asia, and indigenous peoples of the Americas. D5'6 (16189): Mainly in East Asia and Southeast Asia, especially among Chinese people. Generally lower in Siberia, Central Asia, and East India, though the D5a2a2 subclade is prevalent among the Yakuts of Siberia.
 |
The skulls identified as MN00015 and MN00017 in the Museu Nacional/UFRJ in Rio de Janeiro were both from adult male Botocudo individuals from the Rio Doce valley in the state of Minas Gerais, Brazil, as registered by annotations written directly on the outer aspect of their respective parietal bones. According to information of the log book of the museum, it is most likely that these crania arrived there on August 25, 1890. The date of death is not known with certainty, but it is almost certainly the second half of the 19th century.
We first extracted DNA from an intact premolar tooth of skull MN00015. Sequencing of mtDNA identified the following mutations: 73G, 146C, 151T, 6719C, 12239T, 15746G, 16189C, 16217C, 16247G, 16261T. The 9-bp deletion between the COII and tRNA (Lys) genes characteristic of haplogroup B (31) was also present. This set of mutations classified the haplotype as B4a1a, found mainly in Taiwan, Island Southeast Asia and populations on the Pacific Islands. Further analysis identified the mutation 14022G, which classified the sample in haplogroup B4a1a1 (32–34). The presence of the mutation 16247G (HVSI) and 6905A (ancestral allele), further characterized the sequence as belonging to haplogroup B4a1a1a. This haplogroup is found at high frequency in Polynesia, Micronesia, parts of Near Oceania, and Easter Island. As reported previously, 12 of the 14 skulls clearly belonged to the Amerindian haplogroup C1.
The old standard account:
About 12,600 years ago, when ice sheets still covered parts of North America, a baby boy lived, died and was buried in a rocky grave in a field in western Montana. A new whole genome sequencing of this infant — the oldest genome sequence of an American individual — identifies his community as ancestors of Native Americans who live on the continent today. (As always, this type of statement is contradicted elsewhere).
In addition, analysis of the child’s mitochondrial DNA indicated Anzick-1 belonged to what’s known as the D4h3a haplogroup, or lineage. The finding is important — and surprising, according to researchers — because the D4h3a line is considered to be a “founder” lineage, belonging to the first people to arrive in the Americas. Although rare in most Native Americans in the U.S. and Canada today, D4h3a genes are found more commonly in native people of South America, far from the Montana cliff beneath which Anzick-1 was laid to rest.
One of the tricks of the Albino deceivers is to use trick terms. In this case, they use Mal'ta Siberia to imply Mongols like the current people who live there. But in the ancient past, the people of Mal'ta were a Khoisan-like people, as demonstrated by their artifacts and genetics.
Consider these examples:
From Wiki:
In 2014, the autosomal DNA of a male infant from a 12,500-year-old deposit in Montana was sequenced. The DNA was taken from a skeleton referred to as Anzick-1. The skeleton was found in close association with several Clovis artifacts. Comparisons showed strong affinities with DNA from Siberian sites, and virtually ruled out any close affinity of Anzick-1 with European sources (see the "Solutrean hypothesis"). The DNA of the Anzick-1 sample showed strong affinities with sampled Native American populations, which indicated that the samples derive from an ancient population that lived in or near Siberia, the Upper Palaeolithic Mal'ta.
The Study:
Click here for complete pdf file >>
Results: Y-chromosome haplogroup Q-L54*(xM3), Mtdna haplogroup D4h3.
Methods, Results and Conclusions (excerpted):
We analysed 19 different published Siberian populations and found
that allele-frequency-based D-statistics in all cases, with the exclusion
of Naukan, were compatible with a diversification pattern of (Siberian,
(NA, SA)), with no evidence for gene flow into theNAgroup (Supplementar
Information section 15), a pattern that agrees with previously published
results22. This suggests that model (1) is more likely, and that
the structure between NativeAmerican lineages predates the Anzick-1
individual and thus appears to go back to pre-Clovis times.
We used outgroup f3-statistics to evaluate the shared genetic history
between all Native American populations and the Anzick-1 genome,
the 24,000-year-old human sample from Mal’ta, Siberia5 and the 4,000
year-old Saqqaq Palaeo-Eskimo sample from Greenland19. We again
found a closer relationship between Anzick-1 and all NativeAmericans
(Supplementary Information section 15 and Extended Data Fig. 5).
Together with the fact that Anzick-1 shows the same relative affinity
to western and eastern Eurasians, this suggests that the gene flow from
the Mal’ta lineage into Native Americans happened before the NA and
SA groups diverged.
Next,we addressed the relationship of theAnzick-1 genome towholegenome sequences from contemporary humans, including two novel genomes from Karitiana and Mayan individuals, and from the ancient Saqqaq sample19.
We assessed the genome-wide genetic affinity of the Anzick-1 individual
to 143 contemporary non-African human populations by computing
outgroup f3-statistics21, which are informative on the amount of
shared genetic drift between an individual and other populations. The
data set included 52 Native American populations, for which genomic
segments derived from recent European and African admixture have
been excluded22.We found that theAnzick-1 individual showed a statistically
significant closer affinity to all 52 Native American groups than
to any extant Eurasian population. Our results are also consistent with previous models derived from
mtDNA, which imply that Native American populations primarily
derive from a single-source population, but that there was a secondary
movement into northern North America29. However, several different
scenarios are compatible with an early divergence of the NA and SA
groups and analyses of more ancient human remains are needed to
further test the findings and interpretations from this single individual
and to elucidate the complex colonization history of the Arctic and
North American populations.
Source: Harvard Medical School July 21, 2015
Summary:
Native Americans living in the Amazon bear an unexpected genetic connection to indigenous people in Australasia, suggesting a previously unknown wave of migration to the Americas thousands of years ago, a new study has found.
The new study, published July 21 in Nature, indicates that there's more to the story. Pontus Skoglund, first author of the paper and a postdoctoral researcher in the Reich lab, was studying genetic data gathered as part of the 2012 study when he noticed a strange similarity between one or two Native American groups in Brazil and indigenous groups in Australia, New Guinea and the Andaman Islands.
The Tupí-speaking Suruí and Karitiana and the Ge-speaking Xavante of the Amazon had a genetic ancestor more closely related to indigenous Australasians than to any other present-day population. This ancestor doesn't appear to have left measurable traces in other Native American groups in South, Central or North America. The genetic markers from this ancestor don't match any population known to have contributed ancestry to Native Americans, and the geographic pattern can't be explained by post-Columbian European, African or Polynesian mixture with Native Americans, the authors said. They believe the ancestry is much older--perhaps as old as the First Americans. In the ensuing millennia, the ancestral group has disappeared. "We've done a lot of sampling in East Asia and nobody looks like this," said Skoglund. "It's an unknown group that doesn't exist anymore."
The team named the mysterious ancestor Population Y, after the Tupí word for ancestor, "Ypykuéra."
Table of Ancient DNA
Motala-12 - Östergötland, Sweden - 7,000 years. Male Y-dna = I-L460, Mtdna = U2e1 Loschbour - Loschbour, Luxembourg - 8,000 years. Male Y-dna = I-L460, Mtdna = U5b1a LBK - Stuttgart, Germany - 7,500 years. Female Mtdna = T2c2 LBK graveyard at Derenburg Meerenstieg II in Germany - 6,000 ya. Palaeo-Eskimo - Qeqertarsuaq, Greenland - 4,000 ya. Male Y-dna = Q1a, Mtdna = D2a1 Tell Halula, Syria. 7,750 B.C. Mtdna = R0, K, L3, U, H, HV, N. Tell Ramad, Syria. 7,000 B.C. Mtdna = R0, K. Wadi el Makkukh, Israel. 4490-4335 B.C. Males
Mtdna = U3a, H, H6. Tell Kurdu Turkey (Ubaid) - 6,000-4,000 B.C. Mtdna = R0, K, L3, U, H, HV, N, H3a. Barcın Turkey - 6500-6200 B.C.
Y-dna = C1a2, I, J2a, H2, G2a2b2a, Menteşe, Turkey - 6400-500 B.C. Y-dna = G2a2a Mtdna = X2m2 Peștera cu Oase, Romania (falsely called the first European), an extinct lineage of Mtdna = N and a form of Y-dna = F exclusive of G, H, and IJ. Kostenki14 European Russia - 38,700-36,200 years. Male Y-dna = C-V199, Mtdna = U2 Ust'-Ishim Ust'-Ishim, Siberia - 45,000 years. Male Y-dna = K-M526, Mtdna = R Khvalynsk II, Volga River, Samara, Russia. 4700-4000 B.C.
Y-dna = R1b1 (M415), R1a1 (M459), Q1a (F2676), Sopka, Russia, 4000-3000 B.C. Mtdna = U2e, C, D, A10, U5a1, U4, Z. La Braña-Arintero - León, Spain 7,000 years. Male Y-dna = C-V183, Mtdna = U5b2c1 El Portalón, Sierra de Atapuerca, Spain. 4960-4829 ya. Y-dna = I2a2a, Mtdna = H3c NE1 - Polgár-Ferenci-hát, Hungary. Female Mtdna = U5b2c 5070-5310 cal BCKO1 Tiszaszőlős-Domaháza, Hungary - 5650-5780 B.C. Male Y-dna = I-L68, Mtdna = R3 NE5 Kompolt - Kigyósér, Hungary - 4990-5210 B.C. Male Y-dna = C-F3393, Mtdna = J1c NE6 Apc-Berekalja I., Hungary - 4990-5210 B.C. Male Y-dna = C-P255, Mtdna = K1a3a3 NE7 Apc-Berekalja I., Hungary - 4990-5210 B.C. Male Y-dna = I-L1228, Mtdna = N1a Kennewick Man Kennewick, Washington state, USA - 8358 years. Male Y-dna = Q-M199 Mtdna = X2a
StudyPopulation Differentiation of Southern Indian Male Lineages Correlates with Agricultural Expansions Predating the Caste System in
India Genotyped 1,680 Y chromosomes representing 12 tribal and 19 non-tribal (caste) endogamous populations from the predominantly Dravidian-speaking Tamil Nadu state in the southernmost part of India. Tribes and castes were both characterized by an overwhelming proportion of putatively Indian autochthonous Y-chromosomal haplogroups (H-M69, F-M89, R1a1-M17, L1-M27, R2-M124, and C5-M356; 81% combined) with a shared genetic heritage dating back to the late Pleistocene (10–30 Kya), suggesting that more recent Holocene migrations from western Eurasia contributed <20% of the male lineages. We found strong evidence for genetic structure, associated primarily with the current mode of subsistence. Coalescence analysis suggested that the social stratification was established 4–6 Kya and there was little admixture during the last 3 Kya, implying a minimal genetic impact of the Varna (caste) system from the historically-documented Brahmin migrations into the area. In contrast, the overall Y-chromosomal patterns, the time depth of population diversifications and the period of differentiation were best explained by the emergence of agricultural technology in South Asia. These results highlight the utility of detailed local genetic studies within India, without prior assumptions about the importance of Varna rank status for population grouping, to obtain new insights into the relative influences of past demographic events for the population structure of the whole of modern India. |
 |
 |
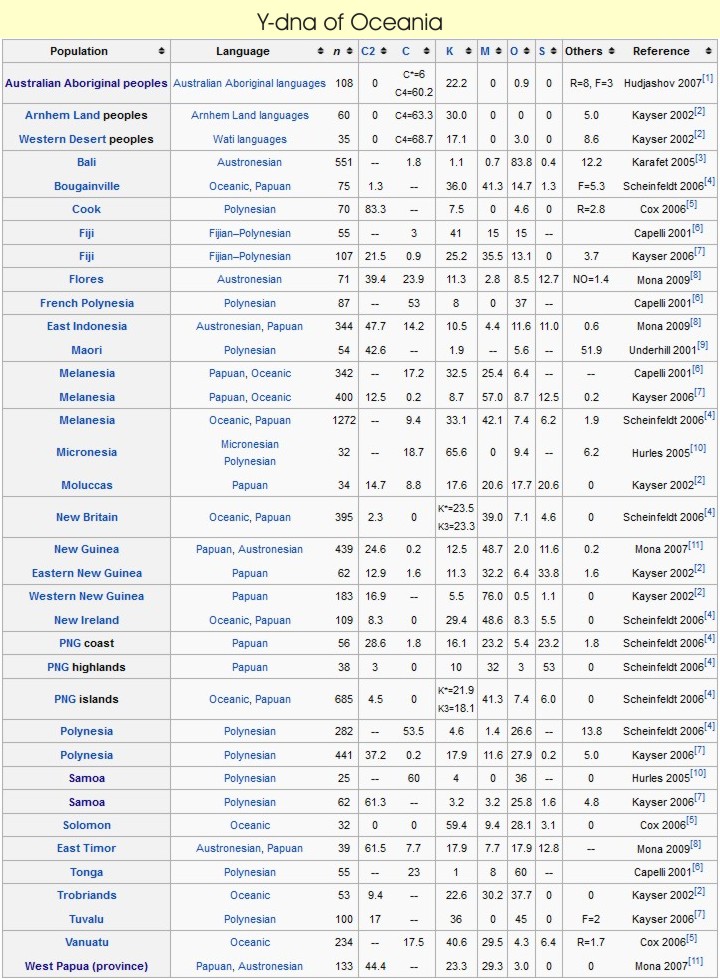 |
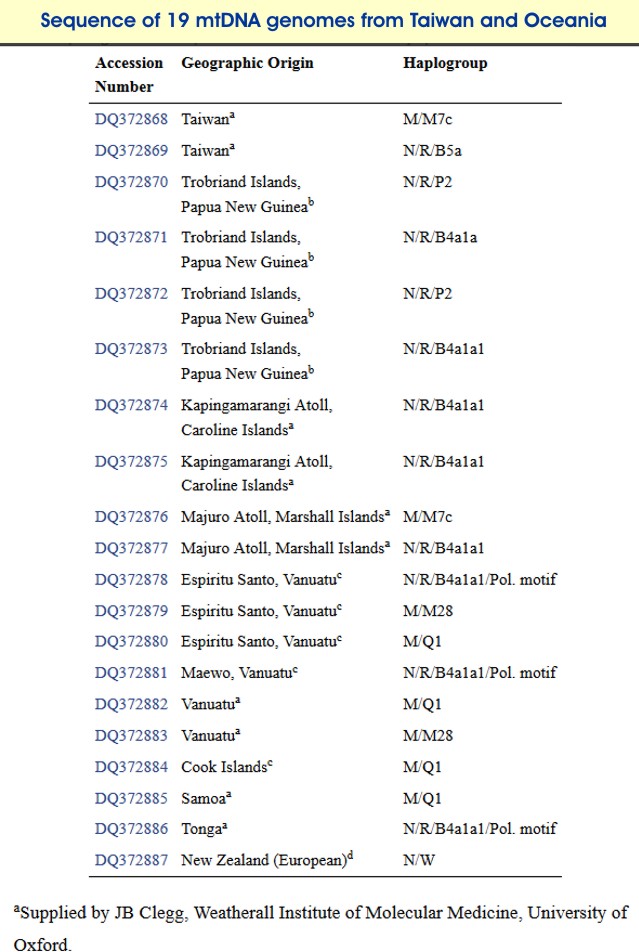 |
 |
 |
 |
 |
 |
 |
 |
 |
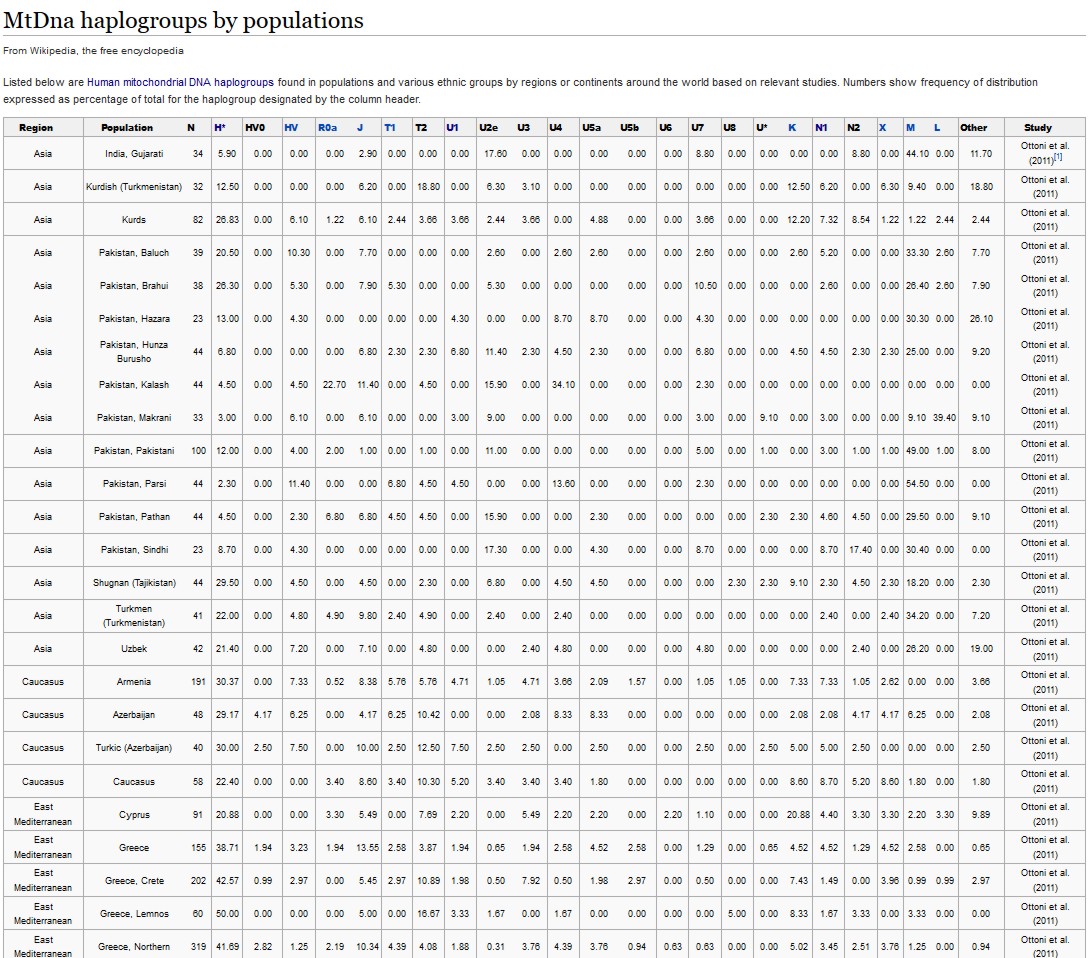 |
 |
 |
African Haplogroup M1
M1 lineages have an African supra-equatorial distribution being mainly present and more diverse in Northeastern and eastern Africa. Occasional occurrences are registered in
West and Northwest Africa Nevertheless, their distribution
is not restricted to Africa, being also relatively
common in the Mediterranean and peaking in the Iberia Peninsula, reaching even the Basque country. M1 has also a well established presence in the Caucasus and the Middle East, ranging from the Arabian Peninsula to Anatolia and
from the Levant to Iran. Central Asian studies have reported its presence as far as Tibet.
African Haplogroup U6
Haplogroup U6 is mostly frequent in
Northwest Africans, being rather frequent in
Algerian Berbers, Moroccans and Mauritanians
but it is also present in Eastern Africans
This
haplogroup is seen as the first return to Africa of
ancient Caucasoid lineages (40-50 kya; Rando
et al. 1998; Maca-Meyer
et al. 2003; Olivieri et
al. (Ha,ha,ha,ha: lying delusional Albinos). joint diffusion of M1 and U6 and arrival in North
Africa in the Early Upper Paleolithic at about 30
kya, likely a human retreat to Africa forced by a
harsh glacial period (González
et al.
, 2007).
The most representative of U6 clades, U6a,
displays increasing frequency and diversity
towards Northwest Africa, supporting the idea
of an autochthonous prehistoric lineage at about
38 kya (Olivieri
et al. 2006). More recent local
dispersal of U6b and U6c sub-clades also paral
lels the distribution of M1b and M1c1 (González et al.
, 2007). One of the most frequent U6a lineages is believed to have started to expand ~11
kya and partially diffused to the Sahel.
African Haplogroup U5
The spatial and temporal overlap for the origin of haplogroups M1 and U6 (Olivieri
et al. 2006) is also true for U5 (Richards
et al. 2000; Achilli
et al. 2005) (Fig.3), which raises the possibility that their molecular ancestors lived in the
same broad geographical area of Southwest Asia (possibly in separate regional enclaves). The main
radiation of U5 took place in Europe, where it
arrived in early Upper Palaeolithic and was likely
among the first AMHs peopling the continent,
most probably from Middle East/Caucasus region
(~40-50 kya; Richards
et al. 2000; Olivieri
et al.
2006). An unexpected finding concerning the
recent U5b1b branch linked the Scandinavian
Saami to the North African Berbers and the sub-Saharan Fulani, not earlier than ~9 kya (Achilli et al. 2005). Together with H1, H3 and V, also
commonly found along the Mediterranean coast
of Africa (Fig.3), these lineages are interpreted as
post-LGM signatures (Achilli
et al. 2004, 2005).
The Franco-Cantabrian refuge area is then seen
as the source of late-glacial surviving lineages carried by the hunter-gatherers that later repopulated most of Europe, and also contributed to the
mtDNA pool of North Africans by crossing the
Strait of Gibraltar (Achilli
et al. 2005). Specific
sub-clades of U5b were found at very low frequencies across sub-Saharan West Africa.
Other N- and R-derived African haplogroups
The presence of haplogroup X in West Eurasia
dates back to pre-Holocene at about 30 kya
(Reidla
et al. 2003), soon after it had diverged
from superhaplogroup N as did its W, N1 (and
I) sister clades. Nowadays, it is present
at low frequencies among West Eurasian, North
Africa and Near East, with specific sub-clades in
Native Americans (Reidla
et al. 2003 and references therein). Clade X1 is largely restricted to
Afro-Asiatic populations in North Africa (particularly Moroccans) and Ethiopians (Kivisild
et al. 2004), Suggesting a diffusion along the
Mediterranean and Red Seas (Reidla
et al. 2003).
The coalescence age of this clade in North Africa
is contemporary to M1 and U6 Paleolithic
mtDNA lineages (Reidla
et al. 2003).
The African maternal pool of those populations inhabiting the Mediterranean coast became
enriched in Eurasian lineages, thanks to several
migratory episodes. The dispersal timeframe of H
lineages (other than H1 and H3) in Egypt is comparable to that of Jordanian H mtDNAs (respectively 21 kya and 23 kya; Roostalu et al. 2007; Cherni
et al. 2009). This drives us to believe in
temporarily coincident events in North Africa
and the Middle East, followed by southward
migrations due to the cold climate of Eurasian
LGM 20 to 14 kya. X2 lineages probably arose
and dispersed around or after the glacial period
not earlier than 25 kya, which is consistent with
their geographic range across all Europe, the
Near East (where it is more diverse), and North
Africa, and wider than that of its X1 sister clade
(Richards
et al. 2000; Reidla
et al. 2003).
Haplogroups J, T1 and R0 have a clear
Middle/Near Eastern origin stated in their age
estimates and declining frequency towards the
southern Caucasus, Near East, Europe and North
and East Africa (Rando
et al. 1998; Krings
et al.
Their spread to
Europe and North Africa is associated with the
Neolithic movements of farming-herding societies 10 to 8 kya. There is nevertheless controversy about the demic input of Near Eastern
Neolithic on the Northwest African autochthonous Capsian Neolithic culture (Newman, 1995)
These maternal lineages may have also diffused
to North Africa in more recent times, perhaps as
lineages of Phoenician traders.
N1 is a minor mtDNA haplogroup that has
been observed at marginal frequencies in European,
Near Eastern, Indian and East African populations (Richards
et al. 2000; Kivisild
et al. 2004),
mainly in Semitic speakers. Although with
much older coalescences in the Near East and
Southwest Asia (Richards
et al. 2000), N1, U
(non-U5 or U6) and W lineages may have been
imported relatively recently, with the expansion
of Semitic languages, at least in the Ethiopian
pool (Kivisild
et al. 2004).
The remaining pool is of West
Eurasian origin, most haplogroups affiliated with
a clear Near Eastern origin. Climatic changes since
the Late Pleistocene to the present day allowed
intense cultural developments and permitted
cross-migrations between Africa and Asia. Among
the first to arrive in the south Mediterranean
basin, as a Near Eastern backflow, are M1 and U6
(and perhaps X1) at about 30 kya (Reidla
et al.
2003; Olivieri
et al.
2006; González
et al. 2007).
Among the later back migrants worth mentioning are H and X2 as post-LGM migrants, and J,
T1 and R0 as Neolithic influences (Richards et
al. 2000; Reidla
et al. 2003). The diffusion of
less frequent N-descendant clades such as N1, W
and I is less understood, but it may trace back to
episode(s) later than their Near Eastern coalescence nearly 25 kya to recent historical times. The
genetic mtDNA profile of Semitic and Cushitic
Afro-Asiatic speakers, inhabitants of Northwest
Africa here considered, are separate from other
population clusters mainly on account of several
Eurasian mtDNA haplogroups, namely JT, H,
V, UK and R0. Other Afro-Asiatic
speakers in Northeast/East Africa, to whom Nilo-Saharan Nubians are in close proximity, cluster on
the responsibility of M1 and minor W, I, N, X
haplogroups.
The Middle East was not the only source
for Eurasian haplotypes of North Africans.
More data has been recently added to support
the hypothesis of an ancient contact between
the Iberian Peninsula and North West Africa
via the Gibraltar Strait, including the analysis
of complete mtDNA sequences, at least in what
concerns post-LGM sub-haplogroups H1, H3,
V and U5b1b (Torroni
et al. 2001a; González
et al. 2003; Achilli
et al.
The trans-Saharan spread of North African
mtDNA legacy is not likely to be a product of
recent gene flow, since a random assortment
of Northwest African mtDNAs would have
also been carried by the migrants. We have to
bear in mind that, even with well documented commercial networks, the Sahara desert seems
to be an important barrier to southward gene
flow of North Africa Eurasiatic haplogroups
(González
et al. 2006).
Evidence of Authentic DNA from Danish Viking Age Skeletons Untouched by Humans for 1,000 Years. Linea Melchior, Toomas Kivisild, Niels Lynnerup - May 2008.
We have assessed mtDNA variation in ten Viking subjects from Galgedil, ca. AD 1,000, in the northern part of Funen, Denmark. All subjects were untouched by humans at the time of sampling. Results - 5 mtdna (H), 1 mtdna (K), 1 mtdna (U5ala), 1 mtdna (I), 1 mtdna (X2C), 1 mtdna (T2).
Several haplogroups that are rare amongst present day Scandinavians were observed in the Galgedil population (three subgroups of Hg H, one subtype of Hg K and one subgroup of Hg X.
 |
| Ref: the last paragraph of the study
For years Albino scientists and researchers have made statements regarding African genetics as if they had actual data to support those statements. They clearly implied that they had done a more-or-less, comprehensive genetic survey of Africa, comparable to their genetic survey of Europe. To those of us who are used to Albino lies and falsifications, who have an understanding of the geographic vastness of Africa, the extremes of terrain, and the uncountable variety of people and cultures, is seemed to be an undoubted falsehood that they could have done such a survey. This study, and that statement, proves that they have not! As a matter of fact: THEY HAVE BARELY SCRATCHED THE GENETIC SURFACE OF AFRICA! And all of those previous statements were merely educated "Guesses", as is much of the data contained on this page. |
| Click here for Realhistoryww Home Page |